Measuring mRNA translation in neuronal processes and somata by tRNA-FRET
- PMID: 31974573
- PMCID: PMC7102941
- DOI: 10.1093/nar/gkaa042
Measuring mRNA translation in neuronal processes and somata by tRNA-FRET
Abstract
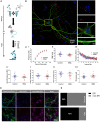
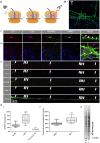
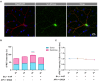
In neurons, the specific spatial and temporal localization of protein synthesis is of great importance for function and survival. Here, we visualized tRNA and protein synthesis events in fixed and live mouse primary cortical culture using fluorescently-labeled tRNAs. We were able to characterize the distribution and transport of tRNAs in different neuronal sub-compartments and to study their association with the ribosome. We found that tRNA mobility in neural processes is lower than in somata and corresponds to patterns of slow transport mechanisms, and that larger tRNA puncta co-localize with translational machinery components and are likely the functional fraction. Furthermore, chemical induction of long-term potentiation (LTP) in culture revealed up-regulation of mRNA translation with a similar effect in dendrites and somata, which appeared to be GluR-dependent 6 h post-activation. Importantly, measurement of protein synthesis in neurons with high resolutions offers new insights into neuronal function in health and disease states.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

