Identification of key genes as potential biomarkers for triple‑negative breast cancer using integrating genomics analysis
- PMID: 31974598
- PMCID: PMC6947886
- DOI: 10.3892/mmr.2019.10867
Identification of key genes as potential biomarkers for triple‑negative breast cancer using integrating genomics analysis
Abstract
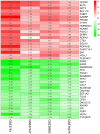
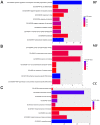
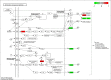
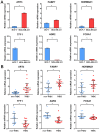
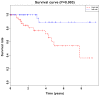
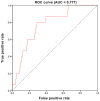
Triple‑negative breast cancer (TNBC) accounts for the worst prognosis of all types of breast cancers due to a high risk of recurrence and a lack of targeted therapeutic options. Extensive effort is required to identify novel targets for TNBC. In the present study, a robust rank aggregation (RRA) analysis based on genome‑wide gene expression datasets involving TNBC patients from the Gene Expression Omnibus (GEO) database was performed to identify key genes associated with TNBC. A total of 194 highly ranked differentially expressed genes (DEGs) were identified in TNBC vs. non‑TNBC. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway (KEGG) enrichment analysis was utilized to explore the biological functions of the identified genes. These DEGs were mainly involved in the biological processes termed positive regulation of transcription from RNA polymerase II promoter, negative regulation of apoptotic process, response to drug, response to estradiol and negative regulation of cell growth. Genes were mainly involved in the KEGG pathway termed estrogen signaling pathway. The aberrant expression of several randomly selected DEGs were further validated in cell lines, clinical tissues and The Cancer Genome Atlas (TCGA) cohort. Furthermore, all the top‑ranked DEGs underwent survival analysis using TCGA database, of which overexpression of 4 genes (FABP7, ART3, CT83, and TTYH1) were positively correlated to the life expectancy (P<0.05) of TNBC patients. In addition, a model consisting of two genes (FABP7 and CT83) was identified to be significantly associated with the overall survival (OS) of TNBC patients by means of Cox regression, Kaplan‑Meier, and receiver operating characteristic (ROC) analyses. In conclusion, the present study identified a number of key genes as potential biomarkers involved in TNBC, which provide novel insights into the tumorigenesis of TNBC at the gene level and may serve as independent prognostic factors for TNBC prognosis.
Keywords: triple-negative breast cancer; robust rank aggregation; biomarker; prognostic risk model; differentially expressed gene.
Figures
Similar articles
-
Identification of differentially expressed genes between triple and non-triple-negative breast cancer using bioinformatics analysis.Breast Cancer. 2019 Nov;26(6):784-791. doi: 10.1007/s12282-019-00988-x. Epub 2019 Jun 13. Breast Cancer. 2019. PMID: 31197620
-
KEGG-expressed genes and pathways in triple negative breast cancer: Protocol for a systematic review and data mining.Medicine (Baltimore). 2020 May;99(18):e19986. doi: 10.1097/MD.0000000000019986. Medicine (Baltimore). 2020. PMID: 32358373 Free PMC article.
-
Highly heterogeneous-related genes of triple-negative breast cancer: potential diagnostic and prognostic biomarkers.BMC Cancer. 2021 May 31;21(1):644. doi: 10.1186/s12885-021-08318-1. BMC Cancer. 2021. PMID: 34053447 Free PMC article.
-
Molecular targets and therapies associated with poor prognosis of triple‑negative breast cancer (Review).Int J Oncol. 2025 Jun;66(6):52. doi: 10.3892/ijo.2025.5758. Epub 2025 May 30. Int J Oncol. 2025. PMID: 40444482 Free PMC article. Review.
-
Next-generation biomarkers for prognostic and potential therapeutic enhancement in Triple negative breast cancer.Crit Rev Oncol Hematol. 2024 Sep;201:104417. doi: 10.1016/j.critrevonc.2024.104417. Epub 2024 Jun 18. Crit Rev Oncol Hematol. 2024. PMID: 38901639 Review.
Cited by
-
The tweety Gene Family: From Embryo to Disease.Front Mol Neurosci. 2021 Jun 28;14:672511. doi: 10.3389/fnmol.2021.672511. eCollection 2021. Front Mol Neurosci. 2021. PMID: 34262434 Free PMC article.
-
Unraveling Biomarker Signatures in Triple-Negative Breast Cancer: A Systematic Review for Targeted Approaches.Int J Mol Sci. 2024 Feb 22;25(5):2559. doi: 10.3390/ijms25052559. Int J Mol Sci. 2024. PMID: 38473804 Free PMC article.
-
Multi-Gene Testing Overview with a Clinical Perspective in Metastatic Triple-Negative Breast Cancer.Int J Mol Sci. 2021 Jul 1;22(13):7154. doi: 10.3390/ijms22137154. Int J Mol Sci. 2021. PMID: 34281208 Free PMC article. Review.
-
Machine learning assisted analysis of breast cancer gene expression profiles reveals novel potential prognostic biomarkers for triple-negative breast cancer.Comput Struct Biotechnol J. 2022 Mar 24;20:1618-1631. doi: 10.1016/j.csbj.2022.03.019. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35465161 Free PMC article.
-
Discovery and Exploration of Small Molecule Binders for CT83: Computational Insights from Homology Modeling, Virtual Screening, MD Simulations, Interaction Fingerprint, and Network Communications.ACS Omega. 2025 May 23;10(22):22884-22908. doi: 10.1021/acsomega.5c00053. eCollection 2025 Jun 10. ACS Omega. 2025. PMID: 40521510 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

