Biosynthesis of Nitrogenase Cofactors
- PMID: 31975585
- PMCID: PMC7318056
- DOI: 10.1021/acs.chemrev.9b00489
Biosynthesis of Nitrogenase Cofactors
Abstract
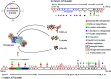
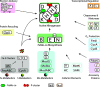
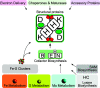
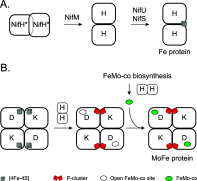
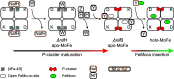
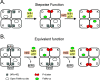
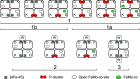
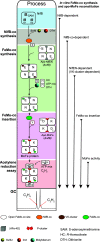
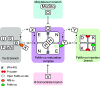
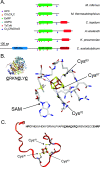
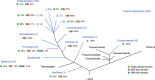
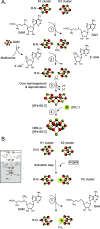
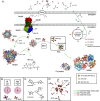
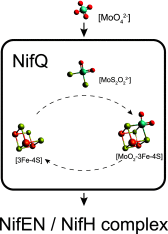
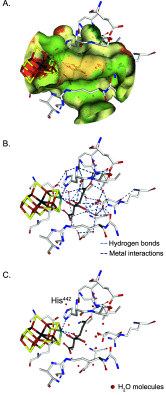
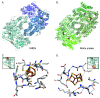
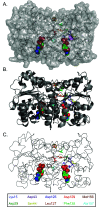
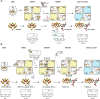
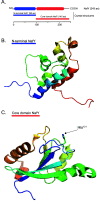
Nitrogenase harbors three distinct metal prosthetic groups that are required for its activity. The simplest one is a [4Fe-4S] cluster located at the Fe protein nitrogenase component. The MoFe protein component carries an [8Fe-7S] group called P-cluster and a [7Fe-9S-C-Mo-R-homocitrate] group called FeMo-co. Formation of nitrogenase metalloclusters requires the participation of the structural nitrogenase components and many accessory proteins, and occurs both in situ, for the P-cluster, and in external assembly sites for FeMo-co. The biosynthesis of FeMo-co is performed stepwise and involves molecular scaffolds, metallochaperones, radical chemistry, and novel and unique biosynthetic intermediates. This review provides a critical overview of discoveries on nitrogenase cofactor structure, function, and activity over the last four decades.
Conflict of interest statement
The authors declare no competing financial interest.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

