Fungal Systematics and Evolution: FUSE 5
- PMID: 31975743
- PMCID: PMC6978154
- DOI: 10.12905/0380.sydowia71-2019-0141
Fungal Systematics and Evolution: FUSE 5
Abstract
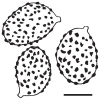
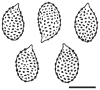
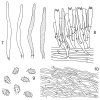
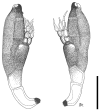
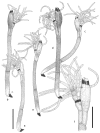
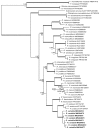
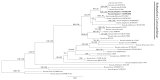
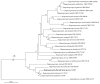
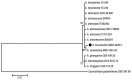
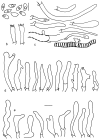
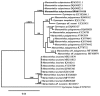
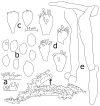
Thirteen new species are formally described: Cortinarius brunneocarpus from Pakistan, C. lilacinoarmillatus from India, Curvularia khuzestanica on Atriplex lentiformis from Iran, Gloeocantharellus neoechinosporus from China, Laboulbenia bernaliana on species of Apenes, Apristus, and Philophuga (Coleoptera, Carabidae) from Nicaragua and Panama, L. oioveliicola on Oiovelia machadoi (Hemiptera, Veliidae) from Brazil, L. termiticola on Macrotermes subhyalinus (Blattodea, Termitidae) from the DR Congo, Pluteus cutefractus from Slovenia, Rhizoglomus variabile from Peru, Russula phloginea from China, Stagonosporopsis flacciduvarum on Vitis vinifera from Italy, Strobilomyces huangshanensis from China, Uromyces klotzschianus on Rumex dentatus subsp. klotzschianus from Pakistan. The following new records are reported: Alternaria calendulae on Calendula officinalis from India; A. tenuissima on apple and quince fruits from Iran; Candelariella oleaginescens from Turkey; Didymella americana and D. calidophila on Vitis vinifera from Italy; Lasiodiplodia theobromae causing tip blight of Dianella tasmanica 'variegata' from India; Marasmiellus subpruinosus from Madeira, Portugal, new for Macaronesia and Africa; Mycena albidolilacea, M. tenuispinosa, and M. xantholeuca from Russia; Neonectria neomacrospora on Madhuca longifolia from India; Nothophoma quercina on Vitis vinifera from Italy; Plagiosphaera immersa on Urtica dioica from Austria; Rinodina sicula from Turkey; Sphaerosporium lignatile from Wisconsin, USA; and Verrucaria murina from Turkey. Multi-locus analysis of ITS, LSU, rpb1, tef1 sequences revealed that P. immersa, commonly classified within Gnomoniaceae (Diaporthales) or as Sordariomycetes incertae sedis, belongs to Magnaporthaceae (Magnaporthales). Analysis of a six-locus Ascomycota-wide dataset including SSU and LSU sequences of S. lignatile revealed that this species, currently in Ascomycota incertae sedis, belongs to Pyronemataceae (Pezizomycetes, Pezizales).
Keywords: 13 new species; 16 new records; Agaricomycetes; Dothideomycetes; Glomeromycota; Laboulbeniomycetes; Magnaporthaceae; Pezizomycetes; Pucciniomycetes; Pyronemataceae; Sordariomycetes; integrative taxonomy.
Figures



















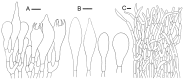



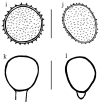
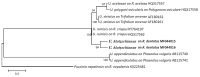

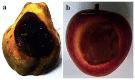














References
-
- Abbasi AM, Khan MA, Ahmad M, Zafar M. Medicinal plant biodiversity of lesser Himalayas-Pakistan. Springer Science & Business Media; Berlin: 2011.
-
- Afshan N, Ishaq A, Niazi AR, Khalid AN. Geographical distribution and host range of genus Uromyces (Pucciniaceae, Uredinales) in Pakistan. International Conference on Chemical, Agricultural and Biological Sciences (CABS-2015); Istanbul (Turkey). 2015. pp. 48–54.
-
- Afshan NS, Khalid AN, Niazi AR, Iqbal SH. New records of Uredinales from Fairy Meadows, Pakistan. Mycotaxon. 2011;115:203–213.
-
- Ahmadpour SA, Mehrabi-Koushki M, Farokhinejad R. Neodidymelliopsis farokhinejadii, a new fungal species from dead branches of trees in Iran. Sydowia. 2017;69:171–182.
Grants and funding
LinkOut - more resources
Full Text Sources
