The complete chloroplast genome of Microcycas calocoma (Miq.) A. DC. (Zamiaceae, Cycadales) and evolution in Cycadales
- PMID: 31976174
- PMCID: PMC6964695
- DOI: 10.7717/peerj.8305
The complete chloroplast genome of Microcycas calocoma (Miq.) A. DC. (Zamiaceae, Cycadales) and evolution in Cycadales
Abstract
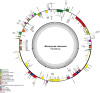
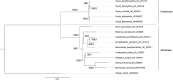
Cycadales is an extant group of seed plants occurring in subtropical and tropical regions comprising putatively three families and 10 genera. At least one complete plastid genome sequence has been reported for all of the 10 genera except Microcycas, making it an ideal plant group to conduct comprehensive plastome comparisons at the genus level. This article reports for the first time the plastid genome of Microcycas calocoma. The plastid genome has a length of 165,688 bp with 134 annotated genes including 86 protein-coding genes, 47 non-coding RNA genes (39 tRNA and eight rRNA) and one pseudogene. Using global sequence variation analysis, the results showed that all cycad genomes share highly similar genomic profiles indicating significant slow evolution and little variation. However, identity matrices coinciding with the inverted repeat regions showed fewer similarities indicating that higher polymorphic events occur at those sites. Conserved non-coding regions also appear to be more divergent whereas variations in the exons were less discernible indicating that the latter comprises more conserved sequences. Phylogenetic analysis using 81 concatenated protein-coding genes of chloroplast (cp) genomes, obtained using maximum likelihood and Bayesian inference with high support values (>70% ML and = 1.0 BPP), confirms that Microcycas is closest to Zamia and forms a monophyletic clade with Ceratozamia and Stangeria. While Stangeria joined the Neotropical cycads Ceratozamia, Zamia and Microcyas, Bowenia grouped with the Southern Hemisphere cycads Encephalartos, Lepidozamia and Macrozamia. All Cycas species formed a distinct clade separated from the other genera. Dioon, on the other hand, was outlying from the rest of Zamiaceae encompassing two major clades-the Southern Hemisphere cycads and the Neotropical cycads. Analysis of the whole cp genomes in phylogeny also supports that the previously recognized family-Stangeriaceae-which contained Bowenia and Stangeria, is not monophyletic. Thus, the cp genome topology obtained in our study is congruent with other molecular phylogenies recognizing only a two-family classification (Cycadaceae and Zamiaceae) within extant Cycadales.
Keywords: Chloroplast genomes; Cycadales; Cycads; Evolution; Microcycas; Phylogeny; Plastids; Zamiaceae; cpDNA.
© 2020 Chang et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25(1):25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
-
- Baldauf SL, Palmer JD. Evolutionary transfer of the chloroplast tufA gene to the nucleus. Nature. 1990;15:262–265. - PubMed
-
- Berry EW. Paleobotany: a sketch of the origin and evolution of floras. In: Walcott CD, editor. Report of the Secretary of the Smithsonian: Institution. Washington, D.C.: Government Printing Office; 1918. pp. 289–407.
LinkOut - more resources
Full Text Sources
Miscellaneous

