Impact of functional studies on exome sequence variant interpretation in early-onset cardiac conduction system diseases
- PMID: 31977013
- PMCID: PMC8453299
- DOI: 10.1093/cvr/cvaa010
Impact of functional studies on exome sequence variant interpretation in early-onset cardiac conduction system diseases
Abstract
Aims: The genetic cause of cardiac conduction system disease (CCSD) has not been fully elucidated. Whole-exome sequencing (WES) can detect various genetic variants; however, the identification of pathogenic variants remains a challenge. We aimed to identify pathogenic or likely pathogenic variants in CCSD patients by using WES and 2015 American College of Medical Genetics and Genomics (ACMG) standards and guidelines as well as evaluating the usefulness of functional studies for determining them.
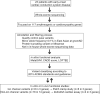
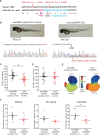
Methods and results: We performed WES of 23 probands diagnosed with early-onset (<65 years) CCSD and analysed 117 genes linked to arrhythmogenic diseases or cardiomyopathies. We focused on rare variants (minor allele frequency < 0.1%) that were absent from population databases. Five probands had protein truncating variants in EMD and LMNA which were classified as 'pathogenic' by 2015 ACMG standards and guidelines. To evaluate the functional changes brought about by these variants, we generated a knock-out zebrafish with CRISPR-mediated insertions or deletions of the EMD or LMNA homologs in zebrafish. The mean heart rate and conduction velocities in the CRISPR/Cas9-injected embryos and F2 generation embryos with homozygous deletions were significantly decreased. Twenty-one variants of uncertain significance were identified in 11 probands. Cellular electrophysiological study and in vivo zebrafish cardiac assay showed that two variants in KCNH2 and SCN5A, four variants in SCN10A, and one variant in MYH6 damaged each gene, which resulted in the change of the clinical significance of them from 'Uncertain significance' to 'Likely pathogenic' in six probands.
Conclusion: Of 23 CCSD probands, we successfully identified pathogenic or likely pathogenic variants in 11 probands (48%). Functional analyses of a cellular electrophysiological study and in vivo zebrafish cardiac assay might be useful for determining the pathogenicity of rare variants in patients with CCSD. SCN10A may be one of the major genes responsible for CCSD.
Keywords: CRISPR/Cas9-mediated gene knock-out in zebrafish; 2015 ACMG standards and guidelines; Cardiac conduction system disease; Cellular electrophysiological study; Whole exome sequencing.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author(s) 2020. For permissions, please email: journals.permissions@oup.com.
Figures





References
-
- Beinart R, Ruskin J, Milan D. The genetics of conduction disease. Heart Fail Clin 2010;6:201–214. - PubMed
-
- Wolf CM, Berul CI. Inherited conduction system abnormalities–one group of diseases, many genes. J Cardiovasc Electrophysiol 2006;17:446–455. - PubMed
-
- Celestino-Soper PB, Doytchinova A, Steiner HA, Uradu A, Lynnes TC, Groh WJ, Miller JM, Lin H, Gao H, Wang Z, Liu Y, Chen PS, Vatta M. Evaluation of the genetic basis of familial aggregation of pacemaker implantation by a large next generation sequencing panel. PLoS One 2015;10:e0143588. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

