AR-induced ZEB1-AS1 represents poor prognosis in cholangiocarcinoma and facilitates tumor stemness, proliferation and invasion through mediating miR-133b/HOXB8
- PMID: 31978895
- PMCID: PMC7053610
- DOI: 10.18632/aging.102680
AR-induced ZEB1-AS1 represents poor prognosis in cholangiocarcinoma and facilitates tumor stemness, proliferation and invasion through mediating miR-133b/HOXB8
Abstract
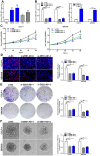
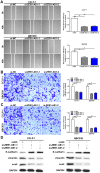
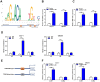
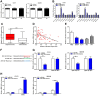
Zinc finger E-box binding homeobox 1 antisense 1 (ZEB1-AS1) has displayed vital regulatory function in various tumors. However, the biological function of ZEB1-AS1 in cholangiocarcinoma (CCA) remains unclear. In this study, we confirmed that ZEB1-AS1 expression was increased in CCA tissues and cells, respectively. Upregulated ZEB1-AS1 was related to lymph node invasion, advanced TNM stage and poor survival of CCA patients. ZEB1-AS1 exhibited high sensitivity and specificity to be an independent poor prognostic factor of patients with CCA. Functionally, knocking down ZEB1-AS1 attenuated tumor cell stemness, restrained cellular viability in vitro and in vivo, and inhibited CCA cell migration and invasion by reversing epithelial-mesenchymal transition. For the mechanism, androgen receptor (AR) directly promoted ZEB1-AS1 expression, and further ZEB1-AS1 increased oncogene homeobox B8 (HOXB8) by sponging miR-133b. In addition, malignant phenotypes of CCA promoted by ZEB1-AS1 dysregulation were rescued separately through interfering miR-133b and HOXB8, suggesting AR/ZEB1-AS1/miR-133b/HOXB8 exerted crucial functions in tumorigenesis and progression of CCA.
Keywords: HOXB8; ZEB1-AS1; cholangiocarcinoma; miR-133b; prognosis.
Conflict of interest statement
Figures




References
-
- Khan SA, Davidson BR, Goldin RD, Heaton N, Karani J, Pereira SP, Rosenberg WM, Tait P, Taylor-Robinson SD, Thillainayagam AV, Thomas HC, Wasan H, and British Society of Gastroenterology. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut. 2012; 61:1657–69. 10.1136/gutjnl-2011-301748 - DOI - PubMed
-
- Sasaki H, Murakami Y, Uemura K, Sudo T, Hashimoto Y, Kondo N, Sueda T. Concurrent analysis of human equilibrative nucleoside transporter 1 and ribonucleotide reductase subunit 1 expression increases predictive value for prognosis in cholangiocarcinoma patients treated with adjuvant gemcitabine-based chemotherapy. Br J Cancer. 2014; 111:1275–84. 10.1038/bjc.2014.399 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

