Conformational changes upon gating of KirBac1.1 into an open-activated state revealed by solid-state NMR and functional assays
- PMID: 31980523
- PMCID: PMC7022178
- DOI: 10.1073/pnas.1915010117
Conformational changes upon gating of KirBac1.1 into an open-activated state revealed by solid-state NMR and functional assays
Abstract
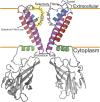
The conformational changes required for activation and K+ conduction in inward-rectifier K+ (Kir) channels are still debated. These structural changes are brought about by lipid binding. It is unclear how this process relates to fast gating or if the intracellular and extracellular regions of the protein are coupled. Here, we examine the structural details of KirBac1.1 reconstituted into both POPC and an activating lipid mixture of 3:2 POPC:POPG (wt/wt). KirBac1.1 is a prokaryotic Kir channel that shares homology with human Kir channels. We establish that KirBac1.1 is in a constitutively active state in POPC:POPG bilayers through the use of real-time fluorescence quenching assays and Förster resonance energy transfer (FRET) distance measurements. Multidimensional solid-state NMR (SSNMR) spectroscopy experiments reveal two different conformers within the transmembrane regions of the protein in this activating lipid environment, which are distinct from the conformation of the channel in POPC bilayers. The differences between these three distinct channel states highlight conformational changes associated with an open activation gate and suggest a unique allosteric pathway that ties the selectivity filter to the activation gate through interactions between both transmembrane helices, the turret, selectivity filter loop, and the pore helix. We also identify specific residues involved in this conformational exchange that are highly conserved among human Kir channels.
Keywords: allostery; lipid activation; membrane protein; potassium channel; solid-state NMR.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Clarke O. B., et al. , Domain reorientation and rotation of an intracellular assembly regulate conduction in Kir potassium channels. Cell 141, 1018–1029 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

