Ancestry-agnostic estimation of DNA sample contamination from sequence reads
- PMID: 31980570
- PMCID: PMC7050530
- DOI: 10.1101/gr.246934.118
Ancestry-agnostic estimation of DNA sample contamination from sequence reads
Abstract
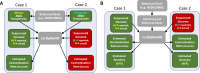
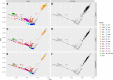
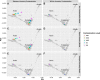
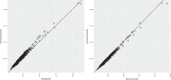
Detecting and estimating DNA sample contamination are important steps to ensure high-quality genotype calls and reliable downstream analysis. Existing methods rely on population allele frequency information for accurate estimation of contamination rates. Correctly specifying population allele frequencies for each individual in early stage of sequence analysis is impractical or even impossible for large-scale sequencing centers that simultaneously process samples from multiple studies across diverse populations. On the other hand, incorrectly specified allele frequencies may result in substantial bias in estimated contamination rates. For example, we observed that existing methods often fail to identify 10% contaminated samples at a typical 3% contamination exclusion threshold when genetic ancestry is misspecified. Such an incomplete screening of contaminated samples substantially inflates the estimated rate of genotyping errors even in deeply sequenced genomes and exomes. We propose a robust statistical method that accurately estimates DNA contamination and is agnostic to genetic ancestry of the intended or contaminating sample. Our method integrates the estimation of genetic ancestry and DNA contamination in a unified likelihood framework by leveraging individual-specific allele frequencies projected from reference genotypes onto principal component coordinates. Our method can also be used for estimating genetic ancestries, similar to LASER or TRACE, but simultaneously accounting for potential contamination. We demonstrate that our method robustly estimates contamination rates and genetic ancestries across populations and contamination scenarios. We further demonstrate that, in the presence of contamination, genetic ancestry inference can be substantially biased with existing methods that ignore contamination, while our method corrects for such biases.
© 2020 Zhang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

References
-
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Automat Contr 19: 716–723. 10.1109/TAC.1974.1100705 - DOI
-
- Brent RP. 1973. Algorithms for minimization without derivatives. Prentice-Hall, Englewood Cliffs, NJ.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
