Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration
- PMID: 31980634
- PMCID: PMC6981263
- DOI: 10.1038/s41467-019-14258-z
Erythroid differentiation regulator-1 induced by microbiota in early life drives intestinal stem cell proliferation and regeneration
Abstract
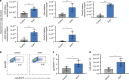
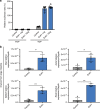
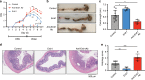
Gut microbiota and their metabolites are instrumental in regulating intestinal homeostasis. However, early-life microbiota associated influences on intestinal development remain incompletely understood. Here we demonstrate that co-housing of germ-free (GF) mice with specific-pathogen free (SPF) mice at weaning (exGF) results in altered intestinal gene expression. Our results reveal that one highly differentially expressed gene, erythroid differentiation regulator-1 (Erdr1), is induced during development in SPF but not GF or exGF mice and localizes to Lgr5+ stem cells and transit amplifying (TA) cells. Erdr1 functions to induce Wnt signaling in epithelial cells, increase Lgr5+ stem cell expansion, and promote intestinal organoid growth. Additionally, Erdr1 accelerates scratch-wound closure in vitro, increases Lgr5+ intestinal stem cell regeneration following radiation-induced injury in vivo, and enhances recovery from dextran sodium sulfate (DSS)-induced colonic damage. Collectively, our findings indicate that early-life microbiota controls Erdr1-mediated intestinal epithelial proliferation and regeneration in response to mucosal damage.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

