Multiple drug binding modes in Mycobacterium tuberculosis CYP51B1
- PMID: 31982812
- PMCID: PMC7092363
- DOI: 10.1016/j.jinorgbio.2020.110994
Multiple drug binding modes in Mycobacterium tuberculosis CYP51B1
Abstract
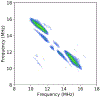
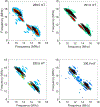
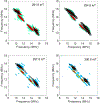
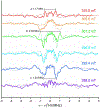
The Mycobacterium tuberculosis (Mtb) genome encodes 20 different cytochrome P450 enzymes (CYPs), many of which serve essential biosynthetic roles. CYP51B1, the Mtb version of eukaryotic sterol demethylase, remains a potential therapeutic target. The binding of three drug fragments containing nitrogen heterocycles to CYP51B1 is studied here by continuous wave (CW) and pulsed electron paramagnetic resonance (EPR) techniques to determine how each drug fragment binds to the heme active-site. All three drug fragments form a mixture of complexes, some of which retain the axial water ligand from the resting state. Hyperfine sublevel correlation spectroscopy (HYSCORE) and electron-nuclear double resonance spectroscopy (ENDOR) observe protons of the axial water and on the drug fragments that reveal drug binding modes. Binding in CYP51B1 is complicated by the presence of multiple binding modes that coexist in the same solution. These results aid our understanding of CYP-inhibitor interactions and will help guide future inhibitor design.
Keywords: Cytochrome P450; EPR; HYSCORE; Mycobacterium tuberculosis.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no conflicts of interest.
Figures









References
-
- McLean KJ, Warman AJ, Seward HE, Marshall KR, Girvan HM, Cheesman MR, Waterman MR, Munro AW, Biophysical characterization of the sterol demethylase P450 from Mycobacterium tuberculosis, its cognate ferredoxin, and their interactions, Biochemistry, 45 (2006) 8427–8443. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

