Myosin Sequestration Regulates Sarcomere Function, Cardiomyocyte Energetics, and Metabolism, Informing the Pathogenesis of Hypertrophic Cardiomyopathy
- PMID: 31983222
- PMCID: PMC7077965
- DOI: 10.1161/CIRCULATIONAHA.119.042339
Myosin Sequestration Regulates Sarcomere Function, Cardiomyocyte Energetics, and Metabolism, Informing the Pathogenesis of Hypertrophic Cardiomyopathy
Erratum in
-
Correction to: Myosin Sequestration Regulates Sarcomere Function, Cardiomyocyte Energetics, and Metabolism, Informing the Pathogenesis of Hypertrophic Cardiomyopathy.Circulation. 2020 Mar 10;141(10):e645. doi: 10.1161/CIR.0000000000000765. Epub 2020 Mar 9. Circulation. 2020. PMID: 32150472 Free PMC article. No abstract available.
Abstract
Background: Hypertrophic cardiomyopathy (HCM) is caused by pathogenic variants in sarcomere protein genes that evoke hypercontractility, poor relaxation, and increased energy consumption by the heart and increased patient risks for arrhythmias and heart failure. Recent studies show that pathogenic missense variants in myosin, the molecular motor of the sarcomere, are clustered in residues that participate in dynamic conformational states of sarcomere proteins. We hypothesized that these conformations are essential to adapt contractile output for energy conservation and that pathophysiology of HCM results from destabilization of these conformations.
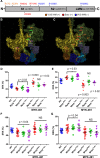
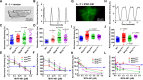
Methods: We assayed myosin ATP binding to define the proportion of myosins in the super relaxed state (SRX) conformation or the disordered relaxed state (DRX) conformation in healthy rodent and human hearts, at baseline and in response to reduced hemodynamic demands of hibernation or pathogenic HCM variants. To determine the relationships between myosin conformations, sarcomere function, and cell biology, we assessed contractility, relaxation, and cardiomyocyte morphology and metabolism, with and without an allosteric modulator of myosin ATPase activity. We then tested whether the positions of myosin variants of unknown clinical significance that were identified in patients with HCM, predicted functional consequences and associations with heart failure and arrhythmias.
Results: Myosins undergo physiological shifts between the SRX conformation that maximizes energy conservation and the DRX conformation that enables cross-bridge formation with greater ATP consumption. Systemic hemodynamic requirements, pharmacological modulators of myosin, and pathogenic myosin missense mutations influenced the proportions of these conformations. Hibernation increased the proportion of myosins in the SRX conformation, whereas pathogenic variants destabilized these and increased the proportion of myosins in the DRX conformation, which enhanced cardiomyocyte contractility, but impaired relaxation and evoked hypertrophic remodeling with increased energetic stress. Using structural locations to stratify variants of unknown clinical significance, we showed that the variants that destabilized myosin conformations were associated with higher rates of heart failure and arrhythmias in patients with HCM.
Conclusions: Myosin conformations establish work-energy equipoise that is essential for life-long cellular homeostasis and heart function. Destabilization of myosin energy-conserving states promotes contractile abnormalities, morphological and metabolic remodeling, and adverse clinical outcomes in patients with HCM. Therapeutic restabilization corrects cellular contractile and metabolic phenotypes and may limit these adverse clinical outcomes in patients with HCM.
Keywords: cardiomyopathy, hypertrophic; cardiovascular physiological phenomena; myosins; sarcomeres.
Figures




References
-
- Barry WH, Bridge JH. Intracellular calcium homeostasis in cardiac myocytes. Circulation. 1993;87:1806–1815. doi: 10.1161/01.cir.87.6.1806. - PubMed
-
- Layland J, Solaro RJ, Shah AM. Regulation of cardiac contractile function by troponin I phosphorylation. Cardiovasc Res. 2005;66:12–21. doi: 10.1016/j.cardiores.2004.12.022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F31 HL149334/HL/NHLBI NIH HHS/United States
- CH/1992001/6764/BHF_/British Heart Foundation/United Kingdom
- RG/18/9/33887/BHF_/British Heart Foundation/United Kingdom
- UL1 TR001863/TR/NCATS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 HL080494/HL/NHLBI NIH HHS/United States
- P50 HL112349/HL/NHLBI NIH HHS/United States
- U01 HL098166/HL/NHLBI NIH HHS/United States
- U01 HL117006/HL/NHLBI NIH HHS/United States
- RG/12/16/29939/BHF_/British Heart Foundation/United Kingdom
- 206466/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- R01 HL084553/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

