Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics
- PMID: 31984744
- PMCID: PMC7127945
- DOI: 10.1021/acs.jproteome.9b00640
Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics
Abstract
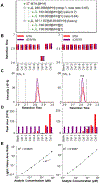
Vendor-independent software tools for quantification of small molecules and metabolites are lacking, especially for targeted analysis workflows. Skyline is a freely available, open-source software tool for targeted quantitative mass spectrometry method development and data processing with a 10 year history supporting six major instrument vendors. Designed initially for proteomics analysis, we describe the expansion of Skyline to data for small molecule analysis, including selected reaction monitoring, high-resolution mass spectrometry, and calibrated quantification. This fundamental expansion of Skyline from a peptide-sequence-centric tool to a molecule-centric tool makes it agnostic to the source of the molecule while retaining Skyline features critical for workflows in both peptide and more general biomolecular research. The data visualization and interrogation features already available in Skyline, such as peak picking, chromatographic alignment, and transition selection, have been adapted to support small molecule data, including metabolomics. Herein, we explain the conceptual workflow for small molecule analysis using Skyline, demonstrate Skyline performance benchmarked against a comparable instrument vendor software tool, and present additional real-world applications. Further, we include step-by-step instructions on using Skyline for small molecule quantitative method development and data analysis on data acquired with a variety of mass spectrometers from multiple instrument vendors.
Keywords: Skyline; data analysis; liquid chromatography; metabolite; metabolomics; quantitative; small molecules; tandem mass spectrometry; targeted.
Figures

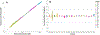

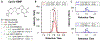
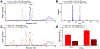
References
-
- Abbatiello SE; Mani DR; Schilling B; Maclean B; Zimmerman LJ; Feng X; Cusack MP; Sedransk N; Hall SC; Addona T; Allen S; Dodder NG; Ghosh M; Held JM; Hedrick V; Inerowicz HD; Jackson A; Keshishian H; Kim JW; Lyssand JS; Riley CP; Rudnick P; Sadowski P; Shaddox K; Smith D; Tomazela D; Wahlander A; Waldemarson S; Whitwell CA; You J; Zhang S; Kinsinger CR; Mesri M; Rodriguez H; Borchers CH; Buck C; Fisher SJ; Gibson BW; Liebler D; Maccoss M; Neubert TA; Paulovich A; Regnier F; Skates SJ; Tempst P; Wang M; Carr SA, Design, implementation and multisite evaluation of a system suitability protocol for the quantitative assessment of instrument performance in liquid chromatography-multiple reaction monitoring-MS (LC-MRM-MS). Molecular & Cellular Proteomics 2013, 12 (9), 2623–2639. - PMC - PubMed
-
- Abbatiello SE; Schilling B; Mani DR; Zimmerman LJ; Hall SC; MacLean B; Albertolle M; Allen S; Burgess M; Cusack MP; Gosh M; Hedrick V; Held JM; Inerowicz HD; Jackson A; Keshishian H; Kinsinger CR; Lyssand J; Makowski L; Mesri M; Rodriguez H; Rudnick P; Sadowski P; Sedransk N; Shaddox K; Skates SJ; Kuhn E; Smith D; Whiteaker JR; Whitwell C; Zhang S; Borchers CH; Fisher SJ; Gibson BW; Liebler DC; MacCoss MJ; Neubert TA; Paulovich AG; Regnier FE; Tempst P; Carr SA, Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Molecular & Cellular Proteomics 2015, 14 (9), 2357–2374. - PMC - PubMed
-
- Schilling B; Rardin MJ; MacLean BX; Zawadzka AM; Frewen BE; Cusack MP; Sorensen DJ; Bereman MS; Jing E; Wu CC; Verdin E; Kahn CR; Maccoss MJ; Gibson BW, Platform-independent and label-free quantitation of proteomic data using MSI extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Molecular & Cellular Proteomics 2012, 11 (5), 202–214. - PMC - PubMed
-
- Rardin MJ, Rapid Assessment of Contaminants and Interferences in Mass Spectrometry Data Using Skyline. Journal of The American Society for Mass Spectrometry 2018, 29 (6), 1327–1330. - PubMed
Publication types
MeSH terms
Grants and funding
- RF1 AG057358/AG/NIA NIH HHS/United States
- R56 AG057895/AG/NIA NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- T32 AG000266/AG/NIA NIH HHS/United States
- R01 GM103551/GM/NIGMS NIH HHS/United States
- K01 AG041211/AG/NIA NIH HHS/United States
- R56 AG063885/AG/NIA NIH HHS/United States
- P30 AG013280/AG/NIA NIH HHS/United States
- P41 GM103533/GM/NIGMS NIH HHS/United States
- P41 GM103493/GM/NIGMS NIH HHS/United States
- RF1 AG057895/AG/NIA NIH HHS/United States
- R01 AG046171/AG/NIA NIH HHS/United States
- R01 AG066184/AG/NIA NIH HHS/United States
- RF1 AG051550/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

