HK97 gp74 Possesses an α-Helical Insertion in the ββα Fold That Affects Its Metal Binding, cos Site Digestion, and In Vivo Activities
- PMID: 31988081
- PMCID: PMC7099138
- DOI: 10.1128/JB.00644-19
HK97 gp74 Possesses an α-Helical Insertion in the ββα Fold That Affects Its Metal Binding, cos Site Digestion, and In Vivo Activities
Abstract
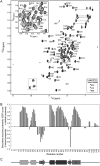
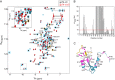
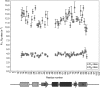
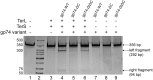
The last gene in the genome of the bacteriophage HK97 encodes gp74, an HNH endonuclease. HNH motifs contain two conserved His residues and an invariant Asn residue, and they adopt a ββα structure. gp74 is essential for phage head morphogenesis, likely because gp74 enhances the specific endonuclease activity of the HK97 terminase complex. Notably, the ability of gp74 to enhance the terminase-mediated cleavage of the phage cos site requires an intact HNH motif in gp74. Mutation of H82, the conserved metal-binding His residue in the HNH motif, to Ala abrogates gp74-mediated stimulation of terminase activity. Here, we present nuclear magnetic resonance (NMR) studies demonstrating that gp74 contains an α-helical insertion in the Ω-loop, which connects the two β-strands of the ββα fold, and a disordered C-terminal tail. NMR data indicate that the Ω-loop insert makes contacts to the ββα fold and influences the ability of gp74 to bind divalent metal ions. Further, the Ω-loop insert and C-terminal tail contribute to gp74-mediated DNA digestion and to gp74 activity in phage morphogenesis. The data presented here enrich our molecular-level understanding of how HNH endonucleases enhance terminase-mediated digestion of the cos site and contribute to the phage replication cycle.IMPORTANCE This study demonstrates that residues outside the canonical ββα fold, namely, the Ω-loop α-helical insert and a disordered C-terminal tail, regulate the activity of the HNH endonuclease gp74. The increased divalent metal ion binding when the Ω-loop insert is removed compared to reduced cos site digestion and phage formation indicates that the Ω-loop insert plays multiple regulatory roles. The data presented here provide insights into the molecular basis of the involvement of HNH proteins in phage DNA packing.
Keywords: HNH endonuclease; NMR spectroscopy; bacteriophage; cos site; divalent metal binding; Ω-loop.
Copyright © 2020 American Society for Microbiology.
Figures

Similar articles
-
The protein gp74 from the bacteriophage HK97 functions as a HNH endonuclease.Protein Sci. 2012 Jun;21(6):809-18. doi: 10.1002/pro.2064. Epub 2012 Apr 23. Protein Sci. 2012. PMID: 22434504 Free PMC article.
-
HNH proteins are a widespread component of phage DNA packaging machines.Proc Natl Acad Sci U S A. 2014 Apr 22;111(16):6022-7. doi: 10.1073/pnas.1320952111. Epub 2014 Apr 7. Proc Natl Acad Sci U S A. 2014. PMID: 24711378 Free PMC article.
-
Strategies for purification of the bacteriophage HK97 small and large terminase subunits that yield pure and homogeneous samples that are functional.Protein Expr Purif. 2019 Aug;160:45-55. doi: 10.1016/j.pep.2019.03.017. Epub 2019 Apr 4. Protein Expr Purif. 2019. PMID: 30954531
-
Sequence-specific DNA nicking endonucleases.Biomol Concepts. 2015 Aug;6(4):253-67. doi: 10.1515/bmc-2015-0016. Biomol Concepts. 2015. PMID: 26352356 Review.
-
Structures, Mechanisms, and Functions of His-Me Finger Nucleases.Trends Biochem Sci. 2020 Nov;45(11):935-946. doi: 10.1016/j.tibs.2020.07.002. Epub 2020 Aug 14. Trends Biochem Sci. 2020. PMID: 32807610 Review.
Cited by
-
Complete Genome Sequence of Stenotrophomonas maltophilia Siphophage Silvanus.Microbiol Resour Announc. 2022 Mar 17;11(3):e0121021. doi: 10.1128/mra.01210-21. Epub 2022 Feb 28. Microbiol Resour Announc. 2022. PMID: 35225669 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

