Whole-genome fingerprint of the DNA methylome during chemically induced differentiation of the human AML cell line HL-60/S4
- PMID: 31988093
- PMCID: PMC7044446
- DOI: 10.1242/bio.044222
Whole-genome fingerprint of the DNA methylome during chemically induced differentiation of the human AML cell line HL-60/S4
Abstract
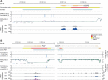
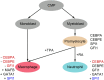
Epigenomic regulation plays a vital role in cell differentiation. The leukemic HL-60/S4 [human myeloid leukemic cell line HL-60/S4 (ATCC CRL-3306)] promyelocytic cell can be easily differentiated from its undifferentiated promyelocyte state into neutrophil- and macrophage-like cell states. In this study, we present the underlying genome and epigenome architecture of HL-60/S4 through its differentiation. We performed whole-genome bisulphite sequencing of HL-60/S4 cells and their differentiated counterparts. With the support of karyotyping, we show that HL-60/S4 maintains a stable genome throughout differentiation. Analysis of differential Cytosine-phosphate-Guanine dinucleotide methylation reveals that most methylation changes occur in the macrophage-like state. Differential methylation of promoters was associated with immune-related terms. Key immune genes, CEBPA, GFI1, MAFB and GATA1 showed differential expression and methylation. However, we observed the strongest enrichment of methylation changes in enhancers and CTCF binding sites, implying that methylation plays a major role in large-scale transcriptional reprogramming and chromatin reorganisation during differentiation. Correlation of differential expression and distal methylation with support from chromatin capture experiments allowed us to identify putative proximal and long-range enhancers for a number of immune cell differentiation genes, including CEBPA and CCNF Integrating expression data, we present a model of HL-60/S4 differentiation in relation to the wider scope of myeloid differentiation.
Keywords: DNA methylation; Differentiation; Epigenomic regulation; HL60; Long range interactions; Promyelocyte.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures



Similar articles
-
Genome wide analysis of acute myeloid leukemia reveal leukemia specific methylome and subtype specific hypomethylation of repeats.PLoS One. 2012;7(3):e33213. doi: 10.1371/journal.pone.0033213. Epub 2012 Mar 29. PLoS One. 2012. PMID: 22479372 Free PMC article.
-
Hypermethylation of the GATA binding protein 4 (GATA4) promoter in Chinese pediatric acute myeloid leukemia.BMC Cancer. 2015 Oct 21;15:756. doi: 10.1186/s12885-015-1760-5. BMC Cancer. 2015. PMID: 26490736 Free PMC article.
-
Cancer-specific changes in DNA methylation reveal aberrant silencing and activation of enhancers in leukemia.Blood. 2017 Feb 16;129(7):e13-e25. doi: 10.1182/blood-2016-07-726877. Epub 2016 Dec 21. Blood. 2017. PMID: 28003272
-
Evolving insights on histone methylome regulation in human acute myeloid leukemia pathogenesis and targeted therapy.Exp Hematol. 2020 Dec;92:19-31. doi: 10.1016/j.exphem.2020.09.189. Epub 2020 Sep 17. Exp Hematol. 2020. PMID: 32950598 Review.
-
Charting the dynamic epigenome during B-cell development.Semin Cancer Biol. 2018 Aug;51:139-148. doi: 10.1016/j.semcancer.2017.08.008. Epub 2017 Aug 26. Semin Cancer Biol. 2018. PMID: 28851627 Review.
Cited by
-
Neutrophil nucleus: shaping the past and the future.J Leukoc Biol. 2023 Nov 24;114(6):585-594. doi: 10.1093/jleuko/qiad084. J Leukoc Biol. 2023. PMID: 37480361 Free PMC article.
-
Linker histone epitopes are hidden by in situ higher-order chromatin structure.Epigenetics Chromatin. 2020 Jun 6;13(1):26. doi: 10.1186/s13072-020-00345-9. Epigenetics Chromatin. 2020. PMID: 32505195 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

