An RNA polymerase ribozyme that synthesizes its own ancestor
- PMID: 31988127
- PMCID: PMC7022166
- DOI: 10.1073/pnas.1914282117
An RNA polymerase ribozyme that synthesizes its own ancestor
Abstract
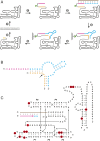
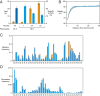
The RNA-based organisms from which modern life is thought to have descended would have depended on an RNA polymerase ribozyme to copy functional RNA molecules, including copying the polymerase itself. Such a polymerase must have been capable of copying structured RNAs with high efficiency and high fidelity to maintain genetic information across successive generations. Here the class I RNA polymerase ribozyme was evolved in vitro for the ability to synthesize functional ribozymes, resulting in the markedly improved ability to synthesize complex RNAs using nucleoside 5'-triphosphate (NTP) substrates. The polymerase is descended from the class I ligase, which contains the same catalytic core as the polymerase. The class I ligase can be synthesized by the improved polymerase as three separate RNA strands that assemble to form a functional ligase. The polymerase also can synthesize the complement of each of these three strands. Despite this remarkable level of activity, only a very small fraction of the assembled ligases retain catalytic activity due to the presence of disabling mutations. Thus, the fidelity of RNA polymerization should be considered a major impediment to the construction of a self-sustained, RNA-based evolving system. The propagation of heritable information requires both efficient and accurate synthesis of genetic molecules, a requirement relevant to both laboratory systems and the early history of life on Earth.
Keywords: RNA enzyme; RNA replication; directed evolution.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

