Transposon Mutagenesis Screen of Klebsiella pneumoniae Identifies Multiple Genes Important for Resisting Antimicrobial Activities of Neutrophils in Mice
- PMID: 31988174
- PMCID: PMC7093148
- DOI: 10.1128/IAI.00034-20
Transposon Mutagenesis Screen of Klebsiella pneumoniae Identifies Multiple Genes Important for Resisting Antimicrobial Activities of Neutrophils in Mice
Abstract
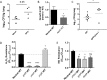
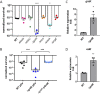
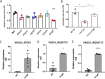
Klebsiella pneumoniae is a Gram-negative bacterial pathogen that causes a range of infections, including pneumonias, urinary tract infections, and septicemia, in otherwise healthy and immunocompromised patients. K. pneumoniae has become an increasing concern due to the rise and spread of antibiotic-resistant and hypervirulent strains. However, its virulence determinants remain understudied. To identify novel K. pneumoniae virulence factors needed to cause pneumonia, a high-throughput screen was performed with an arrayed library of over 13,000 K. pneumoniae transposon insertion mutants in the lungs of wild-type (WT) and neutropenic mice using transposon sequencing (Tn-seq). Insertions in 166 genes resulted in K. pneumoniae mutants that were significantly less fit in the lungs of WT mice than in those of neutropenic mice. Of these, mutants with insertions in 51 genes still had significant defects in neutropenic mice, while mutants with insertions in 52 genes recovered significantly. In vitro screens using a minilibrary of K. pneumoniae transposon mutants identified putative functions for a subset of these genes, including in capsule content and resistance to reactive oxygen and nitrogen species. Lung infections in mice confirmed roles in K. pneumoniae virulence for the ΔdedA, ΔdsbC, ΔgntR, Δwzm-wzt, ΔyaaA, and ΔycgE mutants, all of which were defective in either capsule content or growth in reactive oxygen or nitrogen species. The fitness of the ΔdedA, ΔdsbC, ΔgntR, ΔyaaA, and ΔycgE mutants was higher in neutropenic mouse lungs, indicating that these genes encode proteins that protect K. pneumoniae against neutrophil-related effector functions.
Keywords: Klebsiella; Klebsiella pneumoniae; Tn-seq; dedA; gntR; lung infection; neutropenia; wzm; yaaA; ycgE.
Copyright © 2020 American Society for Microbiology.
Figures



References
-
- Rock C, Thom KA, Masnick M, Johnson JK, Harris AD, Morgan DJ. 2014. Frequency of Klebsiella pneumoniae carbapenemase (KPC)-producing and non-KPC-producing Klebsiella species contamination of healthcare workers and the environment. Infect Control Hosp Epidemiol 35:426–429. doi:10.1086/675598. - DOI - PMC - PubMed
-
- Dao TT, Liebenthal D, Tran TK, Ngoc Thi Vu B, Ngoc Thi Nguyen D, Thi Tran HK, Thi Nguyen CK, Thi Vu HL, Fox A, Horby P, Van Nguyen K, Wertheim H. 2014. Klebsiella pneumoniae oropharyngeal carriage in rural and urban Vietnam and the effect of alcohol consumption. PLoS One 9:e91999. doi:10.1371/journal.pone.0091999. - DOI - PMC - PubMed
-
- Ko WC, Paterson DL, Sagnimeni AJ, Hansen DS, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, McCormack JG, Yu VL. 2002. Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg Infect Dis 8:160–166. doi:10.3201/eid0802.010025. - DOI - PMC - PubMed
-
- Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team . 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi:10.1056/NEJMoa1306801. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

