Swine Influenza A Viruses and the Tangled Relationship with Humans
- PMID: 31988203
- PMCID: PMC7919397
- DOI: 10.1101/cshperspect.a038737
Swine Influenza A Viruses and the Tangled Relationship with Humans
Abstract
Influenza A viruses (IAVs) are the causative agents of one of the most important viral respiratory diseases in pigs and humans. Human and swine IAV are prone to interspecies transmission, leading to regular incursions from human to pig and vice versa. This bidirectional transmission of IAV has heavily influenced the evolutionary history of IAV in both species. Transmission of distinct human seasonal lineages to pigs, followed by sustained within-host transmission and rapid adaptation and evolution, represent a considerable challenge for pig health and production. Consequently, although only subtypes of H1N1, H1N2, and H3N2 are endemic in swine around the world, extensive diversity can be found in the hemagglutinin (HA) and neuraminidase (NA) genes, as well as the remaining six genes. We review the complicated global epidemiology of IAV in swine and the inextricably entangled implications for public health and influenza pandemic planning.
Copyright © 2021 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

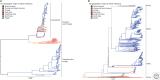
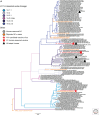



References
-
- Anderson TK, Campbell BA, Nelson MI, Lewis NS, Janas-Martindale A, Killian ML, Vincent AL. 2015. Characterization of co-circulating swine influenza A viruses in North America and the identification of a novel H1 genetic clade with antigenic significance. Virus Res 201: 24–31. 10.1016/j.virusres.2015.02.009 - DOI - PubMed
-
- Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, et al. 2016. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere 1: e00275. 10.1128/mSphere.00275-16 - DOI - PMC - PubMed
-
- Bastien N, Antonishyn NA, Brandt K, Wong CE, Chokani K, Vegh N, Horsman GB, Tyler S, Graham MR, Plummer FA, et al. 2010. Human infection with a triple-reassortant swine influenza A(H1N1) virus containing the hemagglutinin and neuraminidase genes of seasonal influenza virus. J Infect Dis 201: 1178–1182. 10.1086/651507 - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
