Towards a comprehensive catalogue of validated and target-linked human enhancers
- PMID: 31988385
- PMCID: PMC7845138
- DOI: 10.1038/s41576-019-0209-0
Towards a comprehensive catalogue of validated and target-linked human enhancers
Abstract
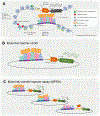
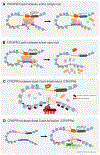
The human gene catalogue is essentially complete, but we lack an equivalently vetted inventory of bona fide human enhancers. Hundreds of thousands of candidate enhancers have been nominated via biochemical annotations; however, only a handful of these have been validated and confidently linked to their target genes. Here we review emerging technologies for discovering, characterizing and validating human enhancers at scale. We furthermore propose a new framework for operationally defining enhancers that accommodates the heterogeneous and complementary results that are emerging from reporter assays, biochemical measurements and CRISPR screens.
Figures



References
-
- Jacob F & Monod J Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol 3, 318–356 (1961). - PubMed
-
- Ptashne M Specific Binding of the λ Phage Repressor to λ DNA. Nature 214, 232–234 (1967). - PubMed
-
- Weintraub H & Groudine M Chromosomal subunits in active genes have an altered conformation. Science 193, 848–856 (1976). - PubMed
-
- Stalder J et al. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell 20, 451–460 (1980). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

