Lentiviral gene therapy for X-linked chronic granulomatous disease
- PMID: 31988463
- PMCID: PMC7115833
- DOI: 10.1038/s41591-019-0735-5
Lentiviral gene therapy for X-linked chronic granulomatous disease
Abstract
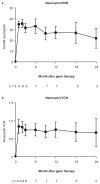
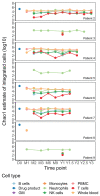
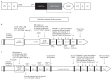
Chronic granulomatous disease (CGD) is a rare inherited disorder of phagocytic cells1,2. We report the initial results of nine severely affected X-linked CGD (X-CGD) patients who received ex vivo autologous CD34+ hematopoietic stem and progenitor cell-based lentiviral gene therapy following myeloablative conditioning in first-in-human studies (trial registry nos. NCT02234934 and NCT01855685). The primary objectives were to assess the safety and evaluate the efficacy and stability of biochemical and functional reconstitution in the progeny of engrafted cells at 12 months. The secondary objectives included the evaluation of augmented immunity against bacterial and fungal infection, as well as assessment of hematopoietic stem cell transduction and engraftment. Two enrolled patients died within 3 months of treatment from pre-existing comorbidities. At 12 months, six of the seven surviving patients demonstrated stable vector copy numbers (0.4-1.8 copies per neutrophil) and the persistence of 16-46% oxidase-positive neutrophils. There was no molecular evidence of either clonal dysregulation or transgene silencing. Surviving patients have had no new CGD-related infections, and six have been able to discontinue CGD-related antibiotic prophylaxis. The primary objective was met in six of the nine patients at 12 months follow-up, suggesting that autologous gene therapy is a promising approach for CGD patients.
Conflict of interest statement
D.B.K. (Kohn), H.L.M., D.A.W. & A.J.T. are Scientific Advisory Board members and H.B.G. is Chief Scientific Officer for Orchard Therapeutics. H.B.G. is an employee and equity/stock holder for Orchard Therapeutics. A.J.T. is an equity/stock holder for Orchard Therapeutics. Orchard Therapeutics has obtained an exclusive option to license from Genethon for the rights and know-how related to the lentiviral vector G1XCGD. C.A.B. and T.P. consult for a sequencing service provider, and C.A.B. consults for Novimmune and SOBI. Eurofins Genomics Sequencing Europe (formerly GATC Biotech AG) is a for-profit company (sequencing service provider). The work performed by Eurofins Genomics Sequencing Europe included in the manuscript is provided to the greater scientific community as a fee for service product. E.C.M. reports Advisory Board attendance for Orchard Therapeutics.
With regards to interests outside of the submitted work, L.D.W. reports grants from the St. Baldrick's Foundation, Damon-Runyon Cancer Research Foundation, and Alex's Lemonade Stand Foundation and personal fees from Magenta Therapeutics. A.J.T. reports Board membership and consultancy with Rocket Pharmaceuticals, Generation Bio, and Board membership with 4BIOCapital. S.-Y.P. reports salary support from Boston Children’s Hospital and a grant from National Institutes of Health. K.L.S. reports personal fees and non-financial support from Orchard Therapeutics, Ltd. E.M.K., G.S., M.A., K.F.B., U.C., S.S.D.R., M.J.D., C.Y.K., D.L.-R., C.R., N.I., K.G., K.S., J.D., E.C.M., D.B.K. (Kuhns), J.G., H.R., J.K.E., G.H., P.E.N., F.D.B., M.G., J.X.-B.D. and A.G. have no competing interests to disclose.
Figures




References
-
- Winkelstein JA, et al. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 2000;79:155–169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

