High-speed motility originates from cooperatively pushing and pulling flagella bundles in bilophotrichous bacteria
- PMID: 31989923
- PMCID: PMC7010408
- DOI: 10.7554/eLife.47551
High-speed motility originates from cooperatively pushing and pulling flagella bundles in bilophotrichous bacteria
Abstract
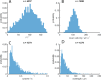
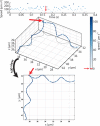
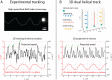
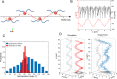
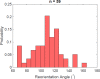
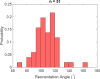
Bacteria propel and change direction by rotating long, helical filaments, called flagella. The number of flagella, their arrangement on the cell body and their sense of rotation hypothetically determine the locomotion characteristics of a species. The movement of the most rapid microorganisms has in particular remained unexplored because of additional experimental limitations. We show that magnetotactic cocci with two flagella bundles on one pole swim faster than 500 µm·s-1 along a double helical path, making them one of the fastest natural microswimmers. We additionally reveal that the cells reorient in less than 5 ms, an order of magnitude faster than reported so far for any other bacteria. Using hydrodynamic modeling, we demonstrate that a mode where a pushing and a pulling bundle cooperate is the only possibility to enable both helical tracks and fast reorientations. The advantage of sheathed flagella bundles is the high rigidity, making high swimming speeds possible.
Keywords: Magnetococcus marinus; dark-field microscopy; magnetotactic bacteria; microswimmer; path tracking; physics of living systems.
© 2020, Bente et al.
Conflict of interest statement
KB, SM, MC, FB, AC, CL, SK, DF No competing interests declared
Figures







References
-
- Bazylinski DA, Williams TJ, Lefèvre CT, Berg RJ, Zhang CL, Bowser SS, Dean AJ, Beveridge TJ. Magnetococcus marinus gen. nov., sp. nov., a marine, magnetotactic bacterium that represents a novel lineage (Magnetococcaceae fam. nov., magnetococcales ord. nov.) at the base of the alphaproteobacteria. International Journal of Systematic and Evolutionary Microbiology. 2013;63:801–808. doi: 10.1099/ijs.0.038927-0. - DOI - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

