Pneumonia recovery reprograms the alveolar macrophage pool
- PMID: 31990682
- PMCID: PMC7101156
- DOI: 10.1172/jci.insight.133042
Pneumonia recovery reprograms the alveolar macrophage pool
Abstract
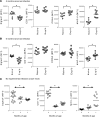
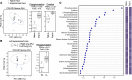
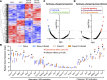
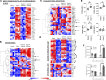
Community-acquired pneumonia is a widespread disease with significant morbidity and mortality. Alveolar macrophages are tissue-resident lung cells that play a crucial role in innate immunity against bacteria that cause pneumonia. We hypothesized that alveolar macrophages display adaptive characteristics after resolution of bacterial pneumonia. We studied mice 1 to 6 months after self-limiting lung infections with Streptococcus pneumoniae, the most common cause of bacterial pneumonia. Alveolar macrophages, but not other myeloid cells, recovered from the lung showed long-term modifications of their surface marker phenotype. The remodeling of alveolar macrophages was (a) long-lasting (still observed 6 months after infection), (b) regionally localized (observed only in the affected lobe after lobar pneumonia), and (c) associated with macrophage-dependent enhanced protection against another pneumococcal serotype. Metabolomic and transcriptomic profiling revealed that alveolar macrophages of mice that recovered from pneumonia had new baseline activities and altered responses to infection that better resembled those of adult humans. The enhanced lung protection after mild and self-limiting bacterial respiratory infections includes a profound remodeling of the alveolar macrophage pool that is long-lasting; compartmentalized; and manifest across surface receptors, metabolites, and both resting and stimulated transcriptomes.
Keywords: Bacterial infections; Innate immunity; Macrophages; Pulmonology.
Conflict of interest statement
Figures





References
-
- Yu H, Wier LM, Elixhauser A. Hospital stays for children, 2009: statistical brief #118. In: Healthcare Cost and Utilization Project (HCUP) Statistical Briefs. Rockville, Maryland, USA: Agency for Healthcare Research and Quality; 2011.
Publication types
MeSH terms
Grants and funding
- F31 HL142199/HL/NHLBI NIH HHS/United States
- K08 HL130582/HL/NHLBI NIH HHS/United States
- R33 HL137081/HL/NHLBI NIH HHS/United States
- R01 HL111459/HL/NHLBI NIH HHS/United States
- F31 HL147397/HL/NHLBI NIH HHS/United States
- R01 HL111449/HL/NHLBI NIH HHS/United States
- UL1 TR001430/TR/NCATS NIH HHS/United States
- R01 GM120060/GM/NIGMS NIH HHS/United States
- R35 HL135756/HL/NHLBI NIH HHS/United States
- R01 AI115053/AI/NIAID NIH HHS/United States
- T32 HL007035/HL/NHLBI NIH HHS/United States
- R01 HL136725/HL/NHLBI NIH HHS/United States
- L30 HL138777/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

