Novel Genetic Variants of ALG6 and GALNTL4 of the Glycosylation Pathway Predict Cutaneous Melanoma-Specific Survival
- PMID: 31991610
- PMCID: PMC7072252
- DOI: 10.3390/cancers12020288
Novel Genetic Variants of ALG6 and GALNTL4 of the Glycosylation Pathway Predict Cutaneous Melanoma-Specific Survival
Abstract
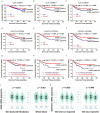
Because aberrant glycosylation is known to play a role in the progression of melanoma, we hypothesize that genetic variants of glycosylation pathway genes are associated with the survival of cutaneous melanoma (CM) patients. To test this hypothesis, we used a Cox proportional hazards regression model in a single-locus analysis to evaluate associations between 34,096 genetic variants of 227 glycosylation pathway genes and CM disease-specific survival (CMSS) using genotyping data from two previously published genome-wide association studies. The discovery dataset included 858 CM patients with 95 deaths from The University of Texas MD Anderson Cancer Center, and the replication dataset included 409 CM patients with 48 deaths from Harvard University nurse/physician cohorts. In the multivariable Cox regression analysis, we found that two novel single-nucleotide polymorphisms (SNPs) (ALG6 rs10889417 G>A and GALNTL4 rs12270446 G>C) predicted CMSS, with an adjusted hazards ratios of 0.60 (95% confidence interval = 0.44-0.83 and p = 0.002) and 0.66 (0.52-0.84 and 0.004), respectively. Subsequent expression quantitative trait loci (eQTL) analysis revealed that ALG6 rs10889417 was associated with mRNA expression levels in the cultured skin fibroblasts and whole blood cells and that GALNTL4 rs12270446 was associated with mRNA expression levels in the skin tissues (all p < 0.05). Our findings suggest that, once validated by other large patient cohorts, these two novel SNPs in the glycosylation pathway genes may be useful prognostic biomarkers for CMSS, likely through modulating their gene expression.
Keywords: cutaneous melanoma; expression quantitative trait loci; glycosylation; single-nucleotide polymorphism; survival analysis.
Conflict of interest statement
The authors have no financial and personal relationships with other people or organizations that could inappropriately influence (bias) their work.
Figures

References
-
- Cancer Stat Facts: Melanoma of the Skin. [(accessed on 11 January 2020)]; Available online: https://seer.cancer.gov/statfacts/html/melan.html.
Grants and funding
- R01 CA100264, 2P50CA093459 and R01CA133996/National Institutes of Health/National Cancer Institute
- P01 CA087969/CA/NCI NIH HHS/United States
- 81573072/the National Natural Science Foundation of China
- NIH CA014236/Duke Cancer Institute as part of the P30 Cancer Centre Support Grant
- R01 CA49449, P01 CA87969, UM1 CA186107 and UM1 CA167552/National Institutes of Health/National Cancer Institute
LinkOut - more resources
Full Text Sources

