Genome Sequence and QTL Analyses Using Backcross Recombinant Inbred Lines (BILs) and BILF1 Lines Uncover Multiple Heterosis-related Loci
- PMID: 31991733
- PMCID: PMC7038202
- DOI: 10.3390/ijms21030780
Genome Sequence and QTL Analyses Using Backcross Recombinant Inbred Lines (BILs) and BILF1 Lines Uncover Multiple Heterosis-related Loci
Abstract
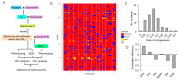
: Heterosis is an interesting topic for both breeders and biologists due to its practical importance and scientific significance. Cultivated rice (Oryza sativa L.) consists of two subspecies, indica and japonica, and hybrid rice is the predominant form of indica rice in China. However, the molecular mechanism underlying heterosis in japonica remains unclear. The present study determined the genome sequence and conducted quantitative trait locus (QTL) analysis using backcross recombinant inbred lines (BILs) and BILF1 lines to uncover the heterosis-related loci for rice yield increase under a japonica genetic background. The BIL population was derived from an admixture variety Habataki and japonica variety Sasanishiki cross to improve the genetic diversity but maintain the genetic background close to japonica. The results showed that heterosis in F1 mainly involved grain number per panicle. The BILF1s showed an increase in grain number per panicle but a decrease in plant height compared with the BILs. Genetic analysis then identified eight QTLs for heterosis in the BILF1s; four QTLs were detected exclusively in the BILF1 population only, presenting a mode of dominance or super-dominance in the heterozygotes. An additional four loci overlapped with QTLs detected in the BIL population, and we found that Grains Height Date 7 (Ghd7) was correlated in days to heading in both BILs and BILF1s. The admixture genetic background of Habataki was also determined by subspecies-specific single nucleotide polymorphisms (SNPs). This investigation highlights the importance of high-throughput sequencing to elucidate the molecular mechanism of heterosis and provides useful germplasms for the application of heterosis in japonica rice production.
Keywords: heterosis; high-throughput sequence; rice; yield components.
Figures





References
-
- Shull G.H. The Genotypes of Maize. Am. Nat. 1911;45:234–252. doi: 10.1086/279207. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

