Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury
- PMID: 31992337
- PMCID: PMC6986156
- DOI: 10.1186/s13041-020-0554-0
Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury
Abstract
Background: N6-methyladenosine (m6A) is the most prevalent post-transcriptional modification of eukaryotic mRNA. It has been reported that there is a stimulus-dependent regulation of m6A in the mammalian central nervous system in response to sensory experience, learning, and injury. The mRNA m6A methylation pattern in rat cortex after traumatic brain injury (TBI) has not been investigated.
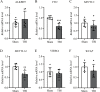
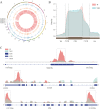
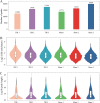
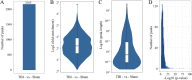
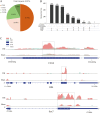
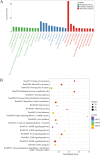
Results: In this study, we conducted a genome-wide profiling of mRNA m6A methylation in rat cortex via methylated RNA immunoprecipitation sequencing (MeRIP-Seq). After TBI, the expressions of METTL14 and FTO were significantly down-regulated in rat cerebral cortex. Using MeRIP-Seq, we identified a total of 2165 significantly changed peaks, of which 1062 were significantly up-regulated and 1103 peaks were significantly down-regulated. These m6A peaks were located across 1850 genes. The analysis of both m6A peaks and mRNA expression revealed that there were 175 mRNA significantly altered methylation and expression levels after TBI. Moreover, it was found that functional FTO is necessary to repair neurological damage caused by TBI but has no effect on the spatial learning and memory abilities of TBI rats by using FTO inhibitor FB23-2.
Conclusion: This study explored the m6A methylation pattern of mRNA after TBI in rat cortex and identified FTO as possible intervention targets in the epigenetic modification of TBI.
Keywords: Epigenetic modification; FTO; Rat cortex; Traumatic brain injury; m6A methylation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

