The genomics of oxidative DNA damage, repair, and resulting mutagenesis
- PMID: 31993111
- PMCID: PMC6974700
- DOI: 10.1016/j.csbj.2019.12.013
The genomics of oxidative DNA damage, repair, and resulting mutagenesis
Abstract
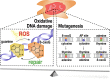
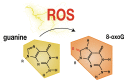
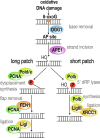
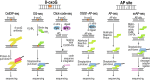
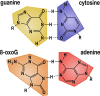
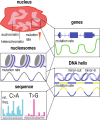
Reactive oxygen species are a constant threat to DNA as they modify bases with the risk of disrupting genome function, inducing genome instability and mutation. Such risks are due to primary oxidative DNA damage and also mediated by the repair process. This leads to a delicate decision process for the cell as to whether to repair a damaged base at a specific genomic location or better leave it unrepaired. Persistent DNA damage can disrupt genome function, but on the other hand it can also contribute to gene regulation by serving as an epigenetic mark. When such processes are out of balance, pathophysiological conditions could get accelerated, because oxidative DNA damage and resulting mutagenic processes are tightly linked to ageing, inflammation, and the development of multiple age-related diseases, such as cancer and neurodegenerative disorders. Recent technological advancements and novel data analysis strategies have revealed that oxidative DNA damage, its repair, and related mutations distribute heterogeneously over the genome at multiple levels of resolution. The involved mechanisms act in the context of genome sequence, in interaction with genome function and chromatin. This review addresses what we currently know about the genome distribution of oxidative DNA damage, repair intermediates, and mutations. It will specifically focus on the various methodologies to measure oxidative DNA damage distribution and discuss the mechanistic conclusions derived from the different approaches. It will also address the consequences of oxidative DNA damage, specifically how it gives rise to mutations, genome instability, and how it can act as an epigenetic mark.
Keywords: 8-oxo-7,8-dihydroguanine; 8-oxoG; AP site; BER; Cancer; DNA damage; Epigenetics; Genomics; Mutagenesis; Oxidative stress; ROS.
© 2020 The Author.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Markkanen E. Not breathing is not an option: How to deal with oxidative DNA damage. DNA Repair (Amst) 2017;59:82–105. - PubMed
-
- van Loon B., Markkanen E., Hübscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst) 2010;9:604–616. - PubMed
-
- Roots R., Okada S. Protection of DNA molecules of cultured mammalian cells from radiation-induced single-strand scissions by various alcohols and SH compounds. Int J Radiat Biol Relat Stud Phys Chem Med. 1972;21:329–342. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
