Age-Related Changes in the Gut Microbiota Modify Brain Lipid Composition
- PMID: 31993375
- PMCID: PMC6970973
- DOI: 10.3389/fcimb.2019.00444
Age-Related Changes in the Gut Microbiota Modify Brain Lipid Composition
Abstract
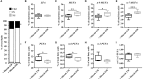
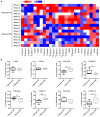
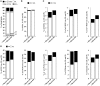
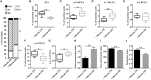
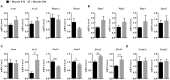
Understanding the molecular mechanisms underlying the changes observed during aging is a prerequisite to design strategies to prevent age-related diseases. Aging is associated with metabolic changes, including alteration in the brain lipid metabolism. These alterations may contribute to the development of pathophysiological conditions. Modifications in the gut microbiota composition are also observed during aging. As communication axes exist between the gut microbiota and the brain and knowing that microbiota influences the host metabolism, we speculated on whether age-associated modifications in the gut microbiota could be involved in the lipid changes observed in aging brain. For that purpose, germ-free mice were colonized by the fecal microbiota of young or old donor mice. Lipid classes and fatty acid profiles were determined in the brain (cortex), plasma and liver by thin-layer chromatography on silica gel-coated quartz rods and gas chromatography. Gut colonization by microbiota of old mice resulted in a significant increase in total monounsaturated fatty acids (MUFA) and a significant decrease in the relative amounts of cholesterol and total polyunsaturated fatty acids (PUFA) in the cortex. Among the eight most represented fatty acids in the cortex, the relative abundances of five (C18:1n-9, C22:6n-3, C20:4n-6, C18:1n-7, and C20:1n-9) were significantly altered in mice inoculated with an aged microbiota. Liquid chromatography analyses revealed that the relative abundance of major species among phosphatidyl and plasmenylcholine (PC 16:0/18:1), phosphatidyl and plasmenylethanolamine (PE 18:0/22:6), lysophosphatidylethanolamine (LPE 22:6) and sphingomyelins (SM d18:1/18:0) were significantly altered in the cortex of mice colonized by the microbiota obtained from aged donors. Transplantation of microbiota from old mice also modified the lipid class and fatty acid content in the liver. Finally, we found that the expression of several genes involved in MUFA and PUFA synthesis (Scd1, Fads1, Fads2, Elovl2, and Elovl5) was dysregulated in mice inoculated with an aged microbiota. In conclusion, our data suggest that changes in gut microbiota that are associated with aging can impact brain and liver lipid metabolisms. Lipid changes induced by an aged microbiota recapitulate some features of aging, thus pointing out the potential role of microbiota alterations in the age-related degradation of the health status.
Keywords: aging; cholesterol; cortex; fatty acid; lipid; liver; microbiota; phospholipid.
Copyright © 2020 Albouery, Buteau, Grégoire, Cherbuy, Pais de Barros, Martine, Chain, Cabaret, Berdeaux, Bron, Acar, Langella and Bringer.
Figures

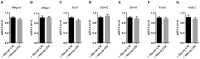
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

