Exome sequencing of bulked segregants identified a novel TaMKK3-A allele linked to the wheat ERA8 ABA-hypersensitive germination phenotype
- PMID: 31993676
- PMCID: PMC7021667
- DOI: 10.1007/s00122-019-03503-0
Exome sequencing of bulked segregants identified a novel TaMKK3-A allele linked to the wheat ERA8 ABA-hypersensitive germination phenotype
Abstract
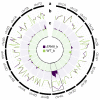
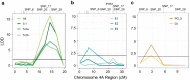
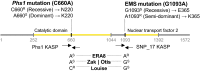
Using bulked segregant analysis of exome sequence, we fine-mapped the ABA-hypersensitive mutant ERA8 in a wheat backcross population to the TaMKK3-A locus of chromosome 4A. Preharvest sprouting (PHS) is the germination of mature grain on the mother plant when it rains before harvest. The ENHANCED RESPONSE TO ABA8 (ERA8) mutant increases seed dormancy and, consequently, PHS tolerance in soft white wheat 'Zak.' ERA8 was mapped to chromosome 4A in a Zak/'ZakERA8' backcross population using bulked segregant analysis of exome sequenced DNA (BSA-exome-seq). ERA8 was fine-mapped relative to mutagen-induced SNPs to a 4.6 Mb region containing 70 genes. In the backcross population, the ERA8 ABA-hypersensitive phenotype was strongly linked to a missense mutation in TaMKK3-A-G1093A (LOD 16.5), a gene associated with natural PHS tolerance in barley and wheat. The map position of ERA8 was confirmed in an 'Otis'/ZakERA8 but not in a 'Louise'/ZakERA8 mapping population. This is likely because Otis carries the same natural PHS susceptible MKK3-A-A660S allele as Zak, whereas Louise carries the PHS-tolerant MKK3-A-C660R allele. Thus, the variation for grain dormancy and PHS tolerance in the Louise/ZakERA8 population likely resulted from segregation of other loci rather than segregation for PHS tolerance at the MKK3 locus. This inadvertent complementation test suggests that the MKK3-A-G1093A mutation causes the ERA8 phenotype. Moreover, MKK3 was a known ABA signaling gene in the 70-gene 4.6 Mb ERA8 interval. None of these 70 genes showed the differential regulation in wild-type Zak versus ERA8 expected of a promoter mutation. Thus, the working model is that the ERA8 phenotype results from the MKK3-A-G1093A mutation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Babraham Bioinformatics (2012) FastQC a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
-
- Bewley JD, Bradford KJ, Hilhorst HWM, Nonogaki H. Seeds. New York: Springer; 2013.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

