Identification of functional domains of the minor fimbrial antigen involved in the interaction of Porphyromonas gingivalis with oral streptococci
- PMID: 31994329
- PMCID: PMC7078856
- DOI: 10.1111/omi.12280
Identification of functional domains of the minor fimbrial antigen involved in the interaction of Porphyromonas gingivalis with oral streptococci
Abstract
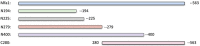
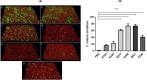
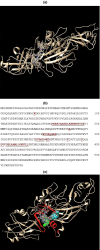
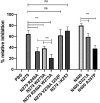
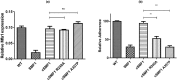
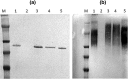
Porphyromonas gingivalis is associated with chronic periodontitis and may initially colonize the oral cavity by adhering to streptococci. Adhesion to streptococci is driven by interaction of the minor fimbrial antigen (Mfa1) with streptococcal antigen I/II. We identified the region of antigen I/II required for this interaction and developed small molecule mimetics that inhibited P. gingivalis adherence. However, the functional motifs of Mfa1 involved in the interaction with antigen I/II remain uncharacterized. A series of N- and C-terminal peptide fragments of Mfa1 were expressed and tested for inhibition of P. gingivalis adherence to S. gordonii. This approach identified residues 225-400 of Mfa1 as essential for P. gingivalis adherence. Using the three-dimensional structure of Mfa1, a putative binding cleft was identified using SiteMap and five small molecule mimetics could dock in this site. Site-specific mutation of residues in the predicted cleft, including R240A, W275A, D321A and A357P inhibited the interaction of Mfa1 with streptococci, whereas mutation of residues not in the predicted cleft (V238A, I252F and ΔK253) had no effect. Complementation of an Mfa1-deficient P. gingivalis strain with wild-type mfa1 restored adherence to streptococci, whereas complementation with full-length mfa1 containing the R240A or A357P mutations did not restore adherence. The mutations did not affect polymerization of Mfa1, suggesting that the complemented strains produced intact minor fimbriae. These results identified specific residues and structural motifs required for the Mfa1-antigen I/II interaction and will facilitate the design of small molecule therapeutics to prevent P. gingivalis colonization of the oral cavity.
Keywords: P. gingivalis; Mfa1; adherence; biofilm; streptococci.
© 2020 The Authors. Molecular Oral Microbiology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain.Infect Immun. 2008 Jul;76(7):3273-80. doi: 10.1128/IAI.00366-08. Epub 2008 May 12. Infect Immun. 2008. PMID: 18474648 Free PMC article.
-
Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates.Microbiology (Reading). 2002 Jun;148(Pt 6):1627-1636. doi: 10.1099/00221287-148-6-1627. Microbiology (Reading). 2002. PMID: 12055284
-
Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation.Infect Immun. 2006 Oct;74(10):5756-62. doi: 10.1128/IAI.00813-06. Infect Immun. 2006. PMID: 16988253 Free PMC article.
-
Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii.Infect Immun. 2005 Jul;73(7):3983-9. doi: 10.1128/IAI.73.7.3983-3989.2005. Infect Immun. 2005. PMID: 15972485 Free PMC article.
-
Porphyromonas gingivalis FimA and Mfa1 fimbriae: Current insights on localization, function, biogenesis, and genotype.Jpn Dent Sci Rev. 2021 Nov;57:190-200. doi: 10.1016/j.jdsr.2021.09.003. Epub 2021 Oct 7. Jpn Dent Sci Rev. 2021. PMID: 34691295 Free PMC article. Review.
Cited by
-
Identification of Small-Molecule Inhibitors Targeting Porphyromonas gingivalis Interspecies Adherence and Determination of Their In Vitro and In Vivo Efficacies.Antimicrob Agents Chemother. 2020 Oct 20;64(11):e00884-20. doi: 10.1128/AAC.00884-20. Print 2020 Oct 20. Antimicrob Agents Chemother. 2020. PMID: 32816725 Free PMC article.
-
An Evolutionary Conservation and Druggability Analysis of Enzymes Belonging to the Bacterial Shikimate Pathway.Antibiotics (Basel). 2022 May 17;11(5):675. doi: 10.3390/antibiotics11050675. Antibiotics (Basel). 2022. PMID: 35625318 Free PMC article.
-
The polymicrobial pathogenicity of Porphyromonas gingivalis.Front Oral Health. 2024 Apr 26;5:1404917. doi: 10.3389/froh.2024.1404917. eCollection 2024. Front Oral Health. 2024. PMID: 38736461 Free PMC article.
-
Effects of Periodontal Treatment in Patients with Periodontitis and Kidney Failure: A Pilot Study.Int J Environ Res Public Health. 2022 Jan 29;19(3):1533. doi: 10.3390/ijerph19031533. Int J Environ Res Public Health. 2022. PMID: 35162556 Free PMC article.
-
Metabolic plasticity enables lifestyle transitions of Porphyromonas gingivalis.NPJ Biofilms Microbiomes. 2021 May 24;7(1):46. doi: 10.1038/s41522-021-00217-4. NPJ Biofilms Microbiomes. 2021. PMID: 34031416 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

