GLIPR1 expression is reduced in multiple myeloma but is not a tumour suppressor in mice
- PMID: 31995627
- PMCID: PMC6988976
- DOI: 10.1371/journal.pone.0228408
GLIPR1 expression is reduced in multiple myeloma but is not a tumour suppressor in mice
Abstract
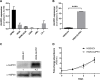
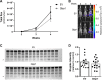
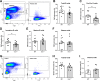
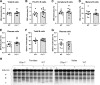
Multiple myeloma, a plasma cell malignancy, is a genetically heterogeneous disease and the genetic factors that contribute to its development and progression remain to be fully elucidated. The tumour suppressor gene GLIPR1 has previously been shown to be deleted in approximately 10% of myeloma patients, to inhibit the development of plasma cell tumours in ageing mice and to have reduced expression levels in the plasma cells of patients with light-chain amyloidosis, a myeloma-related malignancy. Therefore, we hypothesised that GLIPR1 may have tumour suppressor activity in multiple myeloma. In this study, we demonstrate that plasma cell expression of GLIPR1 is reduced in the majority of myeloma patients and Glipr1 expression is lost in the 5TGM1 murine myeloma cell line. However, overexpression of GLIPR1 in a human myeloma cell line did not affect cell proliferation in vitro. Similarly, re-expression of Glipr1 in 5TGM1 cells did not significantly reduce their in vitro proliferation or in vivo growth in C57BL/KaLwRij mice. In addition, using CRISPR-Cas9 genome editing, we generated C57BL/Glipr1-/- mice and showed that loss of Glipr1 in vivo did not affect normal haematopoiesis or the development of monoclonal plasma cell expansions in these mice up to one year of age. Taken together, our results suggest that GLIPR1 is unlikely to be a potent tumour suppressor in multiple myeloma. However, it remains possible that the down-regulation of GLIPR1 may cooperate with other genetic lesions to promote the development of myeloma.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
GLI pathogenesis-related 1 functions as a tumor-suppressor in lung cancer.Mol Cancer. 2016 Mar 18;15:25. doi: 10.1186/s12943-016-0508-4. Mol Cancer. 2016. PMID: 26988096 Free PMC article.
-
Glioma pathogenesis-related protein 1 exerts tumor suppressor activities through proapoptotic reactive oxygen species-c-Jun-NH2 kinase signaling.Cancer Res. 2008 Jan 15;68(2):434-43. doi: 10.1158/0008-5472.CAN-07-2931. Cancer Res. 2008. PMID: 18199537
-
Tetraspanin 7 (TSPAN7) expression is upregulated in multiple myeloma patients and inhibits myeloma tumour development in vivo.Exp Cell Res. 2015 Mar 1;332(1):24-38. doi: 10.1016/j.yexcr.2015.01.006. Epub 2015 Jan 28. Exp Cell Res. 2015. PMID: 25637218
-
Glioma pathogenesis-related protein 1: tumor-suppressor activities and therapeutic potential.Yonsei Med J. 2010 Jul;51(4):479-83. doi: 10.3349/ymj.2010.51.4.479. Yonsei Med J. 2010. PMID: 20499410 Free PMC article. Review.
-
Glioma pathogenesis-related protein 1 performs dual functions in tumor cells.Cancer Gene Ther. 2022 Mar;29(3-4):253-263. doi: 10.1038/s41417-021-00321-9. Epub 2021 Mar 19. Cancer Gene Ther. 2022. PMID: 33742130 Review.
Cited by
-
CAP superfamily proteins in human: a new target for cancer therapy.Med Oncol. 2024 Nov 5;41(12):306. doi: 10.1007/s12032-024-02548-6. Med Oncol. 2024. PMID: 39499355 Review.
-
GliPR1 knockdown by RNA interference exerts anti-glioma effects in vitro and in vivo.J Neurooncol. 2021 May;153(1):23-32. doi: 10.1007/s11060-021-03737-3. Epub 2021 Apr 15. J Neurooncol. 2021. PMID: 33856615 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

