Discovery of a subgenotype of human coronavirus NL63 associated with severe lower respiratory tract infection in China, 2018
- PMID: 31996093
- PMCID: PMC7034077
- DOI: 10.1080/22221751.2020.1717999
Discovery of a subgenotype of human coronavirus NL63 associated with severe lower respiratory tract infection in China, 2018
Abstract
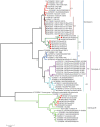
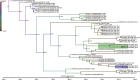
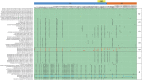
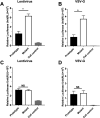
Human coronavirus NL63 (HCoV-NL63) is primarily associated with common cold in children, elderly and immunocompromised individuals. Outbreaks caused by HCoV-NL63 are rare. Here we report a cluster of HCoV-NL63 cases with severe lower respiratory tract infection that arose in Guangzhou, China, in 2018. Twenty-three hospitalized children were confirmed to be HCoV-NL63 positive, and most of whom were hospitalized with severe pneumonia or acute bronchitis. Whole genomes of HCoV-NL63 were obtained using next-generation sequencing. Phylogenetic and single amino acid polymorphism analyses showed that this outbreak was associated with two subgenotypes (C3 and B) of HCoV-NL63. Half of patients were identified to be related to a new subgenotype C3. One unique amino acid mutation at I507 L in spike protein receptor binding domain (RBD) was detected, which segregated this subgenotype C3 from other known subgenotypes. Pseudotyped virus bearing the I507 L mutation in RBD showed enhanced entry into host cells as compared to the prototype virus. This study proved that HCoV-NL63 was undergoing continuous mutation and has the potential to cause severe lower respiratory disease in humans.
Keywords: human coronavirus NL63; new subgenotype; phylogenetic analysis; pneumonia; viral entry; whole-genome sequencing.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
