Genetic background modifies phenotypic severity and longevity in a mouse model of Niemann-Pick disease type C1
- PMID: 31996359
- PMCID: PMC7075069
- DOI: 10.1242/dmm.042614
Genetic background modifies phenotypic severity and longevity in a mouse model of Niemann-Pick disease type C1
Abstract
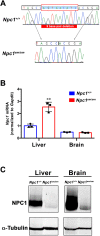
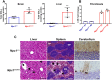
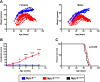
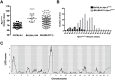
Niemann-Pick disease type C1 (NPC1) is a rare, fatal neurodegenerative disorder characterized by lysosomal accumulation of unesterified cholesterol and glycosphingolipids. These subcellular pathologies lead to phenotypes of hepatosplenomegaly, neurological degeneration and premature death. NPC1 is extremely heterogeneous in the timing of clinical presentation and is associated with a wide spectrum of causative NPC1 mutations. To study the genetic architecture of NPC1, we have generated a new NPC1 mouse model, Npc1em1PavNpc1em1Pav/em1Pav mutants showed notably reduced NPC1 protein compared to controls and displayed the pathological and biochemical hallmarks of NPC1. Interestingly, Npc1em1Pav/em1Pav mutants on a C57BL/6J genetic background showed more severe visceral pathology and a significantly shorter lifespan compared to Npc1em1Pav/em1Pav mutants on a BALB/cJ background, suggesting that strain-specific modifiers contribute to disease severity and survival. QTL analysis for lifespan of 202 backcross N2 mutants on a mixed C57BL/6J and BALB/cJ background detected significant linkage to markers on chromosomes 1 and 7. The discovery of these modifier regions demonstrates that mouse models are powerful tools for analyzing the genetics underlying rare human diseases, which can be used to improve understanding of the variability in NPC1 phenotypes and advance options for patient diagnosis and therapy.This article has an associated First Person interview with the first author of the paper.
Keywords: Genetic modifiers; Mouse models; NPC1; Niemann-Pick disease type C1.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

