Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus
- PMID: 31996437
- PMCID: PMC7081895
- DOI: 10.1128/JVI.00127-20
Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus
Abstract
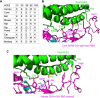
Recently, a novel coronavirus (2019-nCoV) has emerged from Wuhan, China, causing symptoms in humans similar to those caused by severe acute respiratory syndrome coronavirus (SARS-CoV). Since the SARS-CoV outbreak in 2002, extensive structural analyses have revealed key atomic-level interactions between the SARS-CoV spike protein receptor-binding domain (RBD) and its host receptor angiotensin-converting enzyme 2 (ACE2), which regulate both the cross-species and human-to-human transmissions of SARS-CoV. Here, we analyzed the potential receptor usage by 2019-nCoV, based on the rich knowledge about SARS-CoV and the newly released sequence of 2019-nCoV. First, the sequence of 2019-nCoV RBD, including its receptor-binding motif (RBM) that directly contacts ACE2, is similar to that of SARS-CoV, strongly suggesting that 2019-nCoV uses ACE2 as its receptor. Second, several critical residues in 2019-nCoV RBM (particularly Gln493) provide favorable interactions with human ACE2, consistent with 2019-nCoV's capacity for human cell infection. Third, several other critical residues in 2019-nCoV RBM (particularly Asn501) are compatible with, but not ideal for, binding human ACE2, suggesting that 2019-nCoV has acquired some capacity for human-to-human transmission. Last, while phylogenetic analysis indicates a bat origin of 2019-nCoV, 2019-nCoV also potentially recognizes ACE2 from a diversity of animal species (except mice and rats), implicating these animal species as possible intermediate hosts or animal models for 2019-nCoV infections. These analyses provide insights into the receptor usage, cell entry, host cell infectivity and animal origin of 2019-nCoV and may help epidemic surveillance and preventive measures against 2019-nCoV.IMPORTANCE The recent emergence of Wuhan coronavirus (2019-nCoV) puts the world on alert. 2019-nCoV is reminiscent of the SARS-CoV outbreak in 2002 to 2003. Our decade-long structural studies on the receptor recognition by SARS-CoV have identified key interactions between SARS-CoV spike protein and its host receptor angiotensin-converting enzyme 2 (ACE2), which regulate both the cross-species and human-to-human transmissions of SARS-CoV. One of the goals of SARS-CoV research was to build an atomic-level iterative framework of virus-receptor interactions to facilitate epidemic surveillance, predict species-specific receptor usage, and identify potential animal hosts and animal models of viruses. Based on the sequence of 2019-nCoV spike protein, we apply this predictive framework to provide novel insights into the receptor usage and likely host range of 2019-nCoV. This study provides a robust test of this reiterative framework, providing the basic, translational, and public health research communities with predictive insights that may help study and battle this novel 2019-nCoV.
Keywords: 2019-nCoV; SARS coronavirus; angiotensin-converting enzyme 2; animal reservoir; cross-species transmission; human-to-human transmission.
Copyright © 2020 American Society for Microbiology.
Figures



References
-
- Marra MA, Jones SJM, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YSN, Khattra J, Asano JK, Barber SA, Chan SY, Cloutier A, Coughlin SM, Freeman D, Girn N, Griffith OL, Leach SR, Mayo M, McDonald H, Montgomery SB, Pandoh PK, Petrescu AS, Robertson AG, Schein JE, Siddiqui A, Smailus DE, Stott JM, Yang GS, Plummer F, Andonov A, Artsob H, Bastien N, Bernard K, Booth TF, Bowness D, Czub M, Drebot M, Fernando L, Flick R, Garbutt M, Gray M, Grolla A, Jones S, Feldmann H, Meyers A, Kabani A, Li Y, Normand S, Stroher U, Tipples GA, Tyler S, Vogrig R, Ward D, Watson B, Brunham RC, Krajden M, Petric M, Skowronski DM, Upton C, Roper RL. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404. doi: 10.1126/science.1085953. - DOI - PubMed
-
- Peiris JSM, SARS Study Group, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, Nicholls J, Yee WKS, Yan WW, Cheung MT, Cheng VCC, Chan KH, Tsang DNC, Yung RWH, Ng TK, Yuen KY. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. - DOI - PMC - PubMed
-
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JSM, Poon L. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science 302:276–278. doi: 10.1126/science.1087139. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

