Dynamic regulation of anthocyanin biosynthesis at different light intensities by the BT2-TCP46-MYB1 module in apple
- PMID: 31996900
- PMCID: PMC7475178
- DOI: 10.1093/jxb/eraa056
Dynamic regulation of anthocyanin biosynthesis at different light intensities by the BT2-TCP46-MYB1 module in apple
Abstract
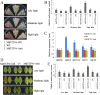
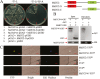
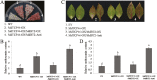
Teosinte branched1/cycloidea/proliferating (TCP) transcription factors play a broad role in plant growth and development, but their involvement in the regulation of anthocyanin biosynthesis is currently unclear. In this study, anthocyanin biosynthesis induced by different light intensities in apple (Malus domestica) was found to be largely dependent on the functions of the MdMYB1 and MdTCP46 transcription factors. The expression of MdTCP46 was responsive to high light intensity, and under these conditions it promoted anthocyanin biosynthesis by direct interactions with MdMYB1 that enhanced the binding of the latter to its target genes. MdTCP46 also interacted with a bric-a-brac/tramtrack/broad (BTB) protein, MdBT2, that is responsive to high light intensity, which ubiquitinated MdTCP46 and mediated its degradation via the 26S proteasome pathway. Our results demonstrate that the dynamic regulatory module MdBT2-MdTCP46-MdMYB1 plays a key role in modulating anthocyanin biosynthesis at different light intensities in apple, and provides new insights into the post-transcriptional regulation of TCP proteins.
Keywords: Malus domestica; Anthocyanin accumulation; TCP transcription factor; apple; high-light intensity; post-transcriptional regulation.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Experimental Biology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures






References
-
- An JP, An XH, Yao JF, Wang XN, You CX, Wang XF, Hao YJ. 2018a BTB protein MdBT2 inhibits anthocyanin and proanthocyanidin biosynthesis by triggering MdMYB9 degradation in apple. Tree Physiology 38, 1578–1587. - PubMed
-
- An JP, Li R, Qu FJ, You CX, Wang XF, Hao YJ. 2018b R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. The Plant Journal 96, 562–577. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

