Fungal Traits Important for Soil Aggregation
- PMID: 31998249
- PMCID: PMC6962133
- DOI: 10.3389/fmicb.2019.02904
Fungal Traits Important for Soil Aggregation
Abstract
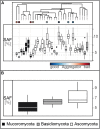
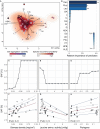
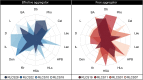
Soil structure, the complex arrangement of soil into aggregates and pore spaces, is a key feature of soils and soil biota. Among them, filamentous saprobic fungi have well-documented effects on soil aggregation. However, it is unclear what properties, or traits, determine the overall positive effect of fungi on soil aggregation. To achieve progress, it would be helpful to systematically investigate a broad suite of fungal species for their trait expression and the relation of these traits to soil aggregation. Here, we apply a trait-based approach to a set of 15 traits measured under standardized conditions on 31 fungal strains including Ascomycota, Basidiomycota, and Mucoromycota, all isolated from the same soil. We find large differences among these fungi in their ability to aggregate soil, including neutral to positive effects, and we document large differences in trait expression among strains. We identify biomass density, i.e., the density with which a mycelium grows (positive effects), leucine aminopeptidase activity (negative effects) and phylogeny as important factors explaining differences in soil aggregate formation (SAF) among fungal strains; importantly, growth rate was not among the important traits. Our results point to a typical suite of traits characterizing fungi that are good soil aggregators, and our findings illustrate the power of employing a trait-based approach to unravel biological mechanisms underpinning soil aggregation. Such an approach could now be extended also to other soil biota groups. In an applied context of restoration and agriculture, such trait information can inform management, for example to prioritize practices that favor the expression of more desirable fungal traits.
Keywords: biomass density; leucine amino peptidases; random forest; saprobic fungi; soil aggregation; traits.
Copyright © 2020 Lehmann, Zheng, Ryo, Soutschek, Roy, Rongstock, Maaß and Rillig.
Figures


References
-
- Aguilar-Trigueros C. A., Hempel S., Powell J. R., Anderson I. C., Antonovics J., Bergmann J., et al. (2015). Branching out: towards a trait-based understanding of fungal ecology. Fungal Biol. Rev. 29 34–41. 10.1016/j.fbr.2015.03.001 - DOI
-
- Andrade-Linares D. R., Veresoglou S. D., Rillig M. C. (2016). Temperature priming and memory in soil filamentous fungi. Fungal Ecol. 21 10–15. 10.1016/j.funeco.2016.02.002 - DOI
-
- Baldrian P., Voriskova J., Dobiasova P., Merhautova V., Lisa L., Valaskova V. (2011). Production of extracellular enzymes and degradation of biopolymers by saprotrophic microfungi from the upper layers of forest soil. Plant Soil 338 111–125. 10.1007/s11104-010-0324-3 - DOI
-
- Bengtsson-Palme J., Ryberg M., Hartmann M., Branco S., Wang Z., Godhe A., et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4 914–919.
LinkOut - more resources
Full Text Sources

