Synthetic Biology Applied to Carbon Conservative and Carbon Dioxide Recycling Pathways
- PMID: 31998710
- PMCID: PMC6966089
- DOI: 10.3389/fbioe.2019.00446
Synthetic Biology Applied to Carbon Conservative and Carbon Dioxide Recycling Pathways
Abstract
The global warming conjugated with our reliance to petrol derived processes and products have raised strong concern about the future of our planet, asking urgently to find sustainable substitute solutions to decrease this reliance and annihilate this climate change mainly due to excess of CO2 emission. In this regard, the exploitation of microorganisms as microbial cell factories able to convert non-edible but renewable carbon sources into biofuels and commodity chemicals appears as an attractive solution. However, there is still a long way to go to make this solution economically viable and to introduce the use of microorganisms as one of the motor of the forthcoming bio-based economy. In this review, we address a scientific issue that must be challenged in order to improve the value of microbial organisms as cell factories. This issue is related to the capability of microbial systems to optimize carbon conservation during their metabolic processes. This initiative, which can be addressed nowadays using the advances in Synthetic Biology, should lead to an increase in products yield per carbon assimilated which is a key performance indice in biotechnological processes, as well as to indirectly contribute to a reduction of CO2 emission.
Keywords: bio-based products; carbon dioxide; chemicals; metabolic engineering; microbial physiology; synthetic biology.
Copyright © 2020 François, Lachaux and Morin.
Figures
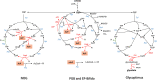
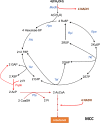
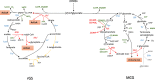
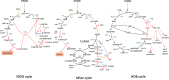
References
-
- Albertyn J., Hohmann S., Thevelein J. M., Prior B. A. (1994). GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14, 4135–4144. 10.1128/MCB.14.6.4135 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

