Dysfunctional polycomb transcriptional repression contributes to lamin A/C-dependent muscular dystrophy
- PMID: 31999646
- PMCID: PMC7190994
- DOI: 10.1172/JCI128161
Dysfunctional polycomb transcriptional repression contributes to lamin A/C-dependent muscular dystrophy
Abstract
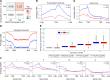
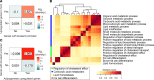
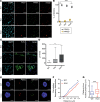
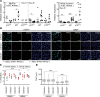
Lamin A is a component of the inner nuclear membrane that, together with epigenetic factors, organizes the genome in higher order structures required for transcriptional control. Mutations in the lamin A/C gene cause several diseases belonging to the class of laminopathies, including muscular dystrophies. Nevertheless, molecular mechanisms involved in the pathogenesis of lamin A-dependent dystrophies are still largely unknown. The polycomb group (PcG) of proteins are epigenetic repressors and lamin A interactors, primarily involved in the maintenance of cell identity. Using a murine model of Emery-Dreifuss muscular dystrophy (EDMD), we show here that lamin A loss deregulated PcG positioning in muscle satellite stem cells, leading to derepression of non-muscle-specific genes and p16INK4a, a senescence driver encoded in the Cdkn2a locus. This aberrant transcriptional program caused impairment in self-renewal, loss of cell identity, and premature exhaustion of the quiescent satellite cell pool. Genetic ablation of the Cdkn2a locus restored muscle stem cell properties in lamin A/C-null dystrophic mice. Our findings establish a direct link between lamin A and PcG epigenetic silencing and indicate that lamin A-dependent muscular dystrophy can be ascribed to intrinsic epigenetic dysfunctions of muscle stem cells.
Keywords: Epigenetics; Mouse stem cells; Muscle Biology; Skeletal muscle; Stem cells.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

