Chromosome Conformation Capture and Beyond: Toward an Integrative View of Chromosome Structure and Function
- PMID: 32001106
- PMCID: PMC7134573
- DOI: 10.1016/j.molcel.2019.12.021
Chromosome Conformation Capture and Beyond: Toward an Integrative View of Chromosome Structure and Function
Abstract
Rapidly developing technologies have recently fueled an exciting era of discovery in the field of chromosome structure and nuclear organization. In addition to chromosome conformation capture (3C) methods, new alternative techniques have emerged to study genome architecture and biological processes in the nucleus, often in single or living cells. This sets an unprecedented stage for exploring the mechanisms that link chromosome structure and biological function. Here we review popular as well as emerging approaches to study chromosome organization, focusing on the contribution of complementary methodologies to our understanding of structures revealed by 3C methods and their biological implications, and discuss the next technical and conceptual frontiers.
Copyright © 2020 Elsevier Inc. All rights reserved.
Figures

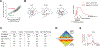

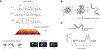
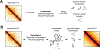
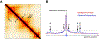
References
-
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, van Lohuizen M, van Steensel B. 2013. Chromatin Position Effects Assayed by Thousands of Reporters Integrated in Parallel. Cell 154: 914–927. - PubMed
-
- Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, Huang B, Lomvardas S, Weiner OD. 2019. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife. https://elifesciences.org/articles/41769 (Accessed September 10, 2019). - PMC - PubMed
-
- Allahyar A, Vermeulen C, Bouwman BAM, Krijger PHL, Verstegen MJAM, Geeven G, van Kranenburg M, Pieterse M, Straver R, Haarhuis JHI, et al. 2018. Enhancer hubs and loop collisions identified from single-allele topologies. Nature Genetics 50: 1151. - PubMed
-
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. 2009. Chromosomal Dynamics at the Shh Locus: Limb Bud-Specific Differential Regulation of Competence and Active Transcription. Developmental Cell 16: 47–57. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

