The DNA repair enzyme MUTYH potentiates cytotoxicity of the alkylating agent MNNG by interacting with abasic sites
- PMID: 32001618
- PMCID: PMC7076218
- DOI: 10.1074/jbc.RA119.010497
The DNA repair enzyme MUTYH potentiates cytotoxicity of the alkylating agent MNNG by interacting with abasic sites
Abstract
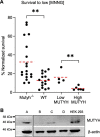
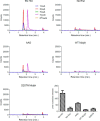
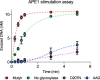
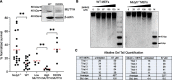
Higher expression of the human DNA repair enzyme MUTYH has previously been shown to be strongly associated with reduced survival in a panel of 24 human lymphoblastoid cell lines exposed to the alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG). The molecular mechanism of MUTYH-enhanced MNNG cytotoxicity is unclear, because MUTYH has a well-established role in the repair of oxidative DNA lesions. Here, we show in mouse embryonic fibroblasts (MEFs) that this MNNG-dependent phenotype does not involve oxidative DNA damage and occurs independently of both O6-methyl guanine adduct cytotoxicity and MUTYH-dependent glycosylase activity. We found that blocking of abasic (AP) sites abolishes higher survival of Mutyh-deficient (Mutyh-/-) MEFs, but this blockade had no additive cytotoxicity in WT MEFs, suggesting the cytotoxicity is due to MUTYH interactions with MNNG-induced AP sites. We found that recombinant mouse MUTYH tightly binds AP sites opposite all four canonical undamaged bases and stimulated apurinic/apyrimidinic endonuclease 1 (APE1)-mediated DNA incision. Consistent with these observations, we found that stable expression of WT, but not catalytically-inactive MUTYH, enhances MNNG cytotoxicity in Mutyh-/- MEFs and that MUTYH expression enhances MNNG-induced genomic strand breaks. Taken together, these results suggest that MUTYH enhances the rapid accumulation of AP-site intermediates by interacting with APE1, implicating MUTYH as a factor that modulates the delicate process of base-excision repair independently of its glycosylase activity.
Keywords: DNA damage; DNA damage response; DNA enzyme; DNA methylation; DNA repair; abasic sites; base excision repair (BER); cancer biology; cancer chemoprevention; mutY homolog (MUTYH).
© 2020 Raetz et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






References
-
- Al-Tassan N., Chmiel N. H., Maynard J., Fleming N., Livingston A. L., Williams G. T., Hodges A. K., Davies D. R., David S. S., Sampson J. R., and Cheadle J. P. (2002) Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 30, 227–232 10.1038/ng828 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

