Two faces of bivalent domain regulate VEGFA responsiveness and angiogenesis
- PMID: 32001672
- PMCID: PMC6992747
- DOI: 10.1038/s41419-020-2228-3
Two faces of bivalent domain regulate VEGFA responsiveness and angiogenesis
Abstract
The bivalent domain (BD) at promoter region is an unique epigenetic feature poised for activation or repression during cell differentiation in embryonic stem cell. However, the function of BDs in already differentiated cells remains exclusive. By profiling the epigenetic landscape of endothelial cells during VEGFA (vascular endothelial growth factor A) stimulation, we discovered that BDs are widespread in endothelial cells and preferentially marked genes responsive to VEGFA. The BDs responsive to VEGFA have more permissive chromatin environment comparing to other BDs. The initial activation of bivalent genes depends on RNAPII pausing release induced by EZH1 rather than removal of H3K27me3. The later suppression of bivalent gene expression depended on KDM5A recruitment by its interaction with PRC2. Importantly, EZH1 promoted both in vitro and in vivo angiogenesis by upregulating EGR3, whereas KDM5A dampened angiogenesis. Collectively, this study demonstrates a novel dual function of BDs in endothelial cells to control VEGF responsiveness and angiogenesis.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures

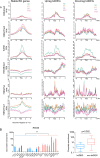
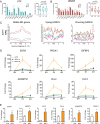

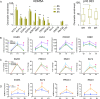

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

