Identification of synergistic drug combinations using breast cancer patient-derived xenografts
- PMID: 32001757
- PMCID: PMC6992640
- DOI: 10.1038/s41598-020-58438-0
Identification of synergistic drug combinations using breast cancer patient-derived xenografts
Abstract
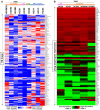
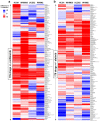
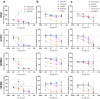
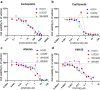
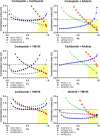
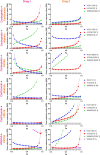
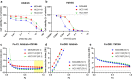
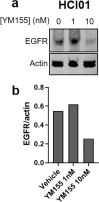
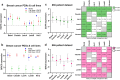
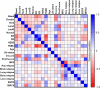
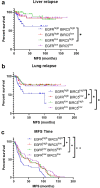
Compared with other breast cancer subtypes, triple-negative breast cancer (TNBC) is associated with relatively poor outcomes due to its metastatic propensity, frequent failure to respond to chemotherapy, and lack of alternative, targeted treatment options, despite decades of major research efforts. Our studies sought to identify promising targeted therapeutic candidates for TNBC through in vitro screening of 1,363 drugs in patient-derived xenograft (PDX) models. Using this approach, we generated a dataset that can be used to assess and compare responses of various breast cancer PDXs to many different drugs. Through a series of further drug screening assays and two-drug combination testing, we identified that the combination of afatinib (epidermal growth factor receptor (EGFR) inhibitor) and YM155 (inhibitor of baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5; survivin) expression) is synergistically cytotoxic across multiple models of basal-like TNBC and reduces PDX mammary tumor growth in vivo. We found that YM155 reduces EGFR expression in TNBC cells, shedding light on its potential mechanism of synergism with afatinib. Both EGFR and BIRC5 are highly expressed in basal-like PDXs, cell lines, and patients, and high expression of both genes reduces metastasis-free survival, suggesting that co-targeting of these proteins holds promise for potential clinical success in TNBC.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Howlader, N. et al. SEERCancer Statistics Review, 1975-2016. National Cancer Institute (2019). Available at, https://seer.cancer.gov/csr/1975_2016/. (Accessed: 27th April 2019).
-
- SEER Cancer Stat Facts: Female Breast Cancer. National Cancer Institute. Available at, https://seer.cancer.gov/statfacts/html/breast.html. (Accessed: 27th April 2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

