Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma
- PMID: 32002016
- PMCID: PMC6986036
- DOI: 10.1186/s12935-020-1113-6
Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) comprises the majority of kidney cancer death worldwide, whose incidence and mortality are not promising. Identifying ideal biomarkers to construct a more accurate prognostic model than conventional clinical parameters is crucial.
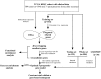
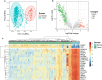
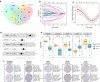
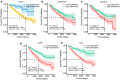
Methods: Raw count of RNA-sequencing data and clinicopathological data were acquired from The Cancer Genome Atlas (TCGA). Tumor samples were divided into two sets. Differentially expressed genes (DEGs) were screened in the whole set and prognosis-related genes were identified from the training set. Their common genes were used in LASSO and best subset regression which were performed to identify the best prognostic 5 genes. The gene-based risk score was developed based on the Cox coefficient of the individual gene. Time-dependent receiver operating characteristic (ROC) and Kaplan-Meier (KM) survival analysis were used to assess its prognostic power. GSE29609 dataset from GEO (Gene Expression Omnibus) database was used to validate the signature. Univariate and multivariate Cox regression were performed to screen independent prognostic parameters to construct a nomogram. The predictive power of the nomogram was revealed by time-dependent ROC curves and the calibration plot and verified in the validation set. Finally, Functional enrichment analysis of DEGs and 5 novel genes were performed to suggest the potential biological pathways.
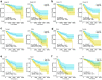
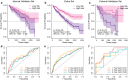
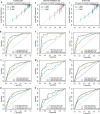
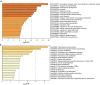
Results: PADI1, ATP6V0D2, DPP6, C9orf135 and PLG were screened to be significantly related to the prognosis of ccRCC patients. The risk score effectively stratified the patients into high-risk group with poor overall survival (OS) based on survival analysis. AJCC-stage, age, recurrence and risk score were regarded as independent prognostic parameters by Cox regression analysis and were used to construct a nomogram. Time-dependent ROC curves showed the nomogram performed best in 1-, 3- and 5-year survival predictions compared with AJCC-stage and risk score in validation sets. The calibration plot showed good agreement of the nomogram between predicted and observed outcomes. Functional enrichment analysis suggested several enriched biological pathways related to cancer.
Conclusions: In our study, we constructed a gene-based model integrating clinical prognostic parameters to predict prognosis of ccRCC well, which might provide a reliable prognosis assessment tool for clinician and aid treatment decision-making in the clinic.
Keywords: Clear cell renal cell carcinoma (ccRCC); Differentially expressed genes (DEGs); GEO; Nomogram; Overall survival (OS); Risk score; TCGA.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

