Are we overestimating risk of enteric pathogen spillover from wild birds to humans?
- PMID: 32003106
- PMCID: PMC7317827
- DOI: 10.1111/brv.12581
Are we overestimating risk of enteric pathogen spillover from wild birds to humans?
Abstract
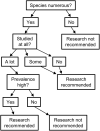
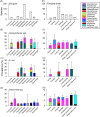
Enteric illnesses remain the second largest source of communicable diseases worldwide, and wild birds are suspected sources for human infection. This has led to efforts to reduce pathogen spillover through deterrence of wildlife and removal of wildlife habitat, particularly within farming systems, which can compromise conservation efforts and the ecosystem services wild birds provide. Further, Salmonella spp. are a significant cause of avian mortality, leading to additional conservation concerns. Despite numerous studies of enteric bacteria in wild birds and policies to discourage birds from food systems, we lack a comprehensive understanding of wild bird involvement in transmission of enteric bacteria to humans. Here, we propose a framework for understanding spillover of enteric pathogens from wild birds to humans, which includes pathogen acquisition, reservoir competence and bacterial shedding, contact with people and food, and pathogen survival in the environment. We place the literature into this framework to identify important knowledge gaps. Second, we conduct a meta-analysis of prevalence data for three human enteric pathogens, Campylobacter spp., E. coli, and Salmonella spp., in 431 North American breeding bird species. Our literature review revealed that only 3% of studies addressed the complete system of pathogen transmission. In our meta-analysis, we found a Campylobacter spp. prevalence of 27% across wild birds, while prevalence estimates of pathogenic E. coli (20%) and Salmonella spp. (6.4%) were lower. There was significant bias in which bird species have been tested, with most studies focusing on a small number of taxa that are common near people (e.g. European starlings Sturnus vulgaris and rock pigeons Columba livia) or commonly in contact with human waste (e.g. gulls). No pathogen prevalence data were available for 65% of North American breeding bird species, including many commonly in contact with humans (e.g. black-billed magpie Pica hudsonia and great blue heron Ardea herodias), and our metadata suggest that some under-studied species, taxonomic groups, and guilds may represent equivalent or greater risk to human infection than heavily studied species. We conclude that current data do not provide sufficient information to determine the likelihood of enteric pathogen spillover from wild birds to humans and thus preclude management solutions. The primary focus in the literature on pathogen prevalence likely overestimates the probability of enteric pathogen spillover from wild birds to humans because a pathogen must survive long enough at an infectious dose and be a strain that is able to colonize humans to cause infection. We propose that future research should focus on the large number of under-studied species commonly in contact with people and food production and demonstrate shedding of bacterial strains pathogenic to humans into the environment where people may contact them. Finally, studies assessing the duration and intensity of bacterial shedding and survival of bacteria in the environment in bird faeces will help provide crucial missing information necessary to calculate spillover probability. Addressing these essential knowledge gaps will support policy to reduce enteric pathogen spillover to humans and enhance bird conservation efforts that are currently undermined by unsupported fears of pathogen spillover from wild birds.
Keywords: Campylobacter spp.; E. coli; Salmonella spp.; agroecology; enteric illness; food safety; wild birds.
© 2020 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
Figures


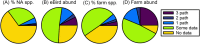

Similar articles
-
Carriage and Subtypes of Foodborne Pathogens Identified in Wild Birds Residing near Agricultural Lands in California: a Repeated Cross-Sectional Study.Appl Environ Microbiol. 2020 Jan 21;86(3):e01678-19. doi: 10.1128/AEM.01678-19. Print 2020 Jan 21. Appl Environ Microbiol. 2020. PMID: 31757824 Free PMC article.
-
Pathogen Presence in European Starlings Inhabiting Commercial Piggeries in South Australia.Avian Dis. 2016 Jun;60(2):430-6. doi: 10.1637/11304-101815-Reg. Avian Dis. 2016. PMID: 27309283
-
A trait-based framework for predicting foodborne pathogen risk from wild birds.Ecol Appl. 2022 Mar;32(2):e2523. doi: 10.1002/eap.2523. Epub 2022 Feb 10. Ecol Appl. 2022. PMID: 34921463
-
Bacterial pathogens in wild birds: a review of the frequency and effects of infection.Biol Rev Camb Philos Soc. 2009 Aug;84(3):349-73. doi: 10.1111/j.1469-185X.2008.00076.x. Epub 2009 Apr 29. Biol Rev Camb Philos Soc. 2009. PMID: 19438430 Review.
-
An annotated checklist of pathogenic microorganisms associated with migratory birds.J Wildl Dis. 2004 Oct;40(4):639-59. doi: 10.7589/0090-3558-40.4.639. J Wildl Dis. 2004. PMID: 15650082 Review.
Cited by
-
Metagenomic analysis of gut microbiome and resistome of Whooper and Black Swans: a one health perspective.BMC Genomics. 2023 Oct 24;24(1):635. doi: 10.1186/s12864-023-09742-2. BMC Genomics. 2023. PMID: 37875797 Free PMC article.
-
Human Campylobacteriosis-A Serious Infectious Threat in a One Health Perspective.Curr Top Microbiol Immunol. 2021;431:1-23. doi: 10.1007/978-3-030-65481-8_1. Curr Top Microbiol Immunol. 2021. PMID: 33620646 Review.
-
Emerging biofilm formation and disinfectant susceptibility of ESBL-producing Klebsiella pneumoniae.Sci Rep. 2025 Jan 10;15(1):1599. doi: 10.1038/s41598-024-84149-x. Sci Rep. 2025. PMID: 39794383 Free PMC article.
-
Quantifying antimicrobial resistance in food-producing animals in North America.Front Microbiol. 2025 May 27;16:1542472. doi: 10.3389/fmicb.2025.1542472. eCollection 2025. Front Microbiol. 2025. PMID: 40497057 Free PMC article.
-
Survey on the Presence of Bacterial and Parasitic Zoonotic Agents in the Feces of Wild Birds.Vet Sci. 2021 Aug 25;8(9):171. doi: 10.3390/vetsci8090171. Vet Sci. 2021. PMID: 34564565 Free PMC article.
References
-
- Abdollahpour, N. , Zendehbad, B. , Alipour, A. & Khayatzadeh, J. (2015). Wild–bird feces as a source of Campylobacter jejuni infection in children's playgrounds in Iran. Food Control 50, 378–381.
-
- * Adesiyun, A. A. , Seepersadsingh, N. , Inder, L. & Caesar, K. (1998). Some bacterial enteropathogens in wildlife and racing pigeons from Trinidad. Journal of Wildlife Diseases 34, 73–80. - PubMed
-
- * Aguirre, A. A. , Quan, T. J. , Cook, R. S. & Mclean, R. G. (1992). Cloacal flora isolated from wild black‐bellied whistling ducks (Dendrocygna autumnalis) in Laguna La Nacha, Mexico. Avian Diseases 36, 459–462. - PubMed
-
- Albuquerque, A. , Cardoso, W. , Teixeira, R. , Lopes, E. , Sales, R. , Horn, R. , Rocha‐E‐Silva, R. , Bezerra, W. & Gomes‐Filho, V. (2013). Dissemination of Salmonella Enterititids by experimentally–infected pigeons. Brazilian Journal of Poultry Science 15, 211–215.
-
- * Alcalá, L. , Alonso, C. A. , Simón, C. , González‐Esteban, C. , Orós, J. , Rezusta, A. , Ortega, C. & Torres, C. (2016). Wild birds, frequent carriers of extended‐spectrum β‐lactamase (ESBL) producing Escherichia coli of CTX‐M and SHV‐12 types. Microbial Ecology 72, 861–869. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

