Host engulfment pathway controls inflammation in inflammatory bowel disease
- PMID: 32003126
- PMCID: PMC7390677
- DOI: 10.1111/febs.15236
Host engulfment pathway controls inflammation in inflammatory bowel disease
Abstract
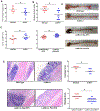
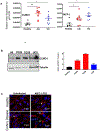
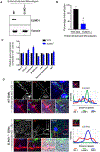
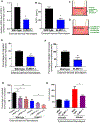
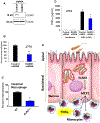
Chronic diseases, including inflammatory bowel disease (IBD) urgently need new biomarkers as a significant proportion of patients, do not respond to current medications. Inflammation is a common factor in these diseases, and microbial sensing in the intestinal tract is critical to initiate the inflammation. We have identified ELMO1 (engulfment and cell motility protein 1) as a microbial sensor in epithelial and phagocytic cells that turns on inflammatory signals. Using a stem cell-based 'gut-in-a-dish' coculture model, we studied the interactions between microbes, epithelium, and monocytes in the context of IBD. To mimic the in vivo cell physiology, enteroid-derived monolayers (EDMs) were generated from the organoids isolated from WT and ELMO1-/- mice and colonic biopsies of IBD patients. The EDMs were infected with the IBD-associated microbes to monitor the inflammatory responses. ELMO1-depleted EDMs displayed a significant reduction in bacterial internalization, a decrease in pro-inflammatory cytokine productions and monocyte recruitment. The expression of ELMO1 is elevated in the colonic epithelium and in the inflammatory infiltrates within the lamina propria of IBD patients where the higher expression is positively correlated with the elevated expression of pro-inflammatory cytokines, MCP-1 and TNF-α. MCP-1 is released from the epithelium and recruits monocytes to the site of inflammation. Once recruited, monocytes require ELMO1 to engulf the bacteria and propagate a robust TNF-α storm. These findings highlight that the dysregulated epithelial ELMO1 → MCP-1 axis can serve as an early biomarker in the diagnostics of IBD and other inflammatory disorders.
Keywords: enteroid; epithelial-immune cell crosstalk; inflammatory bowel disease; microbial sensing and bacterial engulfment; monocyte chemoattractant protein 1.
© 2020 University of California. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
Figures



References
-
- Moustafa A, Li W, Anderson EL, Wong EHM, Dulai PS, Sandborn WJ, Biggs W, Yooseph S, Jones MB, Venter JC, Nelson KE, Chang JT, Telenti A & Boland BS (2018) Genetic risk, dysbiosis, and treatment stratification using host genome and gut microbiome in inflammatory bowel disease, Clin Transl Gastroenterol. 9, e132. - PMC - PubMed
-
- Manichanh C, Borruel N, Casellas F & Guarner F (2012) The gut microbiota in IBD, Nat Rev Gastroenterol Hepatol. 9, 599–608. - PubMed
-
- Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M & Thomas G (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease, Nature. 411, 599–603. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

