The characterization of mobile colistin resistance (mcr) genes among 33 000 Salmonella enterica genomes from routine public health surveillance in England
- PMID: 32003708
- PMCID: PMC7067213
- DOI: 10.1099/mgen.0.000331
The characterization of mobile colistin resistance (mcr) genes among 33 000 Salmonella enterica genomes from routine public health surveillance in England
Abstract
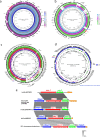
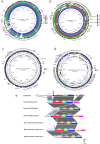
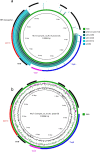
To establish the prevalence of mobile colistin resistance (mcr) genes amongst Salmonella enterica isolates obtained through public health surveillance in England (April 2014 to September 2017), 33 205 S. enterica genome sequences obtained from human, food, animal and environmental isolates were screened for the presence of mcr variants 1 to 8. The mcr-positive genomes were assembled, annotated and characterized according to plasmid type. Nanopore sequencing was performed on six selected isolates with putative novel plasmids, and phylogenetic analysis was used to provide an evolutionary context for the most commonly isolated clones. Fifty-two mcr-positive isolates were identified, of which 32 were positive for mcr-1, 19 for mcr-3 and 1 for mcr-5. The combination of Illumina and Nanopore sequencing identified three novel mcr-3 plasmids and one novel mcr-5 plasmid, as well as the presence of chromosomally integrated mcr-1 and mcr-3. Monophasic S. enterica serovar Typhimurium accounted for 27/52 (52 %) of the mcr-positive isolates, with the majority clustering in clades associated with travel to Southeast Asia. Isolates in these clades were associated with a specific plasmid range and an additional extended-spectrum beta-lactamase genotype. Routine whole-genome sequencing for public health surveillance provides an effective screen for novel and emerging antimicrobial determinants, including mcr. Complementary long-read technologies elucidated the genomic context of resistance determinants, offering insights into plasmid dissemination and linkage to other resistance genes.
Keywords: MCR; Salmonella; WGS; colistin.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Chromosomally and Plasmid-Located mcr in Salmonella from Animals and Food Products in China.Microbiol Spectr. 2022 Dec 21;10(6):e0277322. doi: 10.1128/spectrum.02773-22. Epub 2022 Nov 21. Microbiol Spectr. 2022. PMID: 36409077 Free PMC article.
-
Comparative analysis of an mcr-4 Salmonella enterica subsp. enterica monophasic variant of human and animal origin.J Antimicrob Chemother. 2018 Dec 1;73(12):3332-3335. doi: 10.1093/jac/dky340. J Antimicrob Chemother. 2018. PMID: 30137382
-
Salmonella enterica mcr-1 Positive from Food in Brazil: Detection and Characterization.Foodborne Pathog Dis. 2020 Mar;17(3):202-208. doi: 10.1089/fpd.2019.2700. Epub 2019 Sep 26. Foodborne Pathog Dis. 2020. PMID: 31556704
-
Plasmid-mediated colistin resistance in Latin America and Caribbean: A systematic review.Travel Med Infect Dis. 2019 Sep-Oct;31:101459. doi: 10.1016/j.tmaid.2019.07.015. Epub 2019 Jul 20. Travel Med Infect Dis. 2019. PMID: 31336179
-
Prevalence and Traits of Mobile Colistin Resistance Gene Harbouring Isolates from Different Ecosystems in Africa.Biomed Res Int. 2021 Jan 22;2021:6630379. doi: 10.1155/2021/6630379. eCollection 2021. Biomed Res Int. 2021. PMID: 33553426 Free PMC article. Review.
Cited by
-
Two colistin resistance-producing Aeromonas strains, isolated from coastal waters in Zhejiang, China: characteristics, multi-drug resistance and pathogenicity.Front Microbiol. 2024 Jul 31;15:1401802. doi: 10.3389/fmicb.2024.1401802. eCollection 2024. Front Microbiol. 2024. PMID: 39144207 Free PMC article.
-
Mobile Colistin Resistance (mcr) Gene-Containing Organisms in Poultry Sector in Low- and Middle-Income Countries: Epidemiology, Characteristics, and One Health Control Strategies.Antibiotics (Basel). 2023 Jun 28;12(7):1117. doi: 10.3390/antibiotics12071117. Antibiotics (Basel). 2023. PMID: 37508213 Free PMC article. Review.
-
Evidence of international transmission of mobile colistin resistant monophasic Salmonella Typhimurium ST34.Sci Rep. 2023 May 1;13(1):7080. doi: 10.1038/s41598-023-34242-4. Sci Rep. 2023. PMID: 37127697 Free PMC article.
-
Global epidemiology, genetic environment, risk factors and therapeutic prospects of mcr genes: A current and emerging update.Front Cell Infect Microbiol. 2022 Aug 26;12:941358. doi: 10.3389/fcimb.2022.941358. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36093193 Free PMC article. Review.
-
Characterization of a P1-bacteriophage-like plasmid (phage-plasmid) harbouring bla CTX-M-15 in Salmonella enterica serovar Typhi.Microb Genom. 2022 Dec;8(12):mgen000913. doi: 10.1099/mgen.0.000913. Microb Genom. 2022. PMID: 36748517 Free PMC article.
References
-
- Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar paratyphi B. J Antimicrob Chemother. 2017;72:3317–3324. doi: 10.1093/jac/dkx327. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

