Autophagy-related gene P4HB: a novel diagnosis and prognosis marker for kidney renal clear cell carcinoma
- PMID: 32003756
- PMCID: PMC7053637
- DOI: 10.18632/aging.102715
Autophagy-related gene P4HB: a novel diagnosis and prognosis marker for kidney renal clear cell carcinoma
Abstract
Autophagy can protect cells and organisms from stressors such as nutrient deprivation, and is involved in many pathological processes including human cancer. Therefore, it is necessary to investigate the role of autophagy-related genes (ARGs) in cancer. In this study, we investigated the gene expression of 222 ARGs in 1048 Kidney Renal Clear Cell Carcinoma (KIRC) cases, from 5 independent cohorts. The gene expression of ARGs were first evaluated in the The Cancer Genome Atlas (TCGA) by Recevier Operating Characteristic (ROC) analysis to select potential biomarkers with extremely high ability in KIRC detection (AUC≥0.85 and p<0.0001). Then in silico procedure progressively leads to the selection of two genes in a three rounds of validation performed in four human KIRC-patients datasets including two independent Gene Expression Omnibus (GEO) datasets, Oncomine dataset and Human Protein Atlas dataset. Finally, only P4HB (Prolyl 4-hydroxylase, beta polypeptide) gene was experimentally validated by RT-PCR between control kidney cells and cancer cells. Following univariate and multivariate analyses of TCGA-KIRC clinical data showed that P4HB expression is an independent prognostic indicator of unfavorable overall survival (OS) for KIRC patients. Based on these findings, we proposed that P4HB might be one potential novel KIRC diagnostic and prognostic biomarker at both mRNA and protein levels.
Keywords: KIRC; P4HB; autophagy; diagnostic biomarker; prognostic biomarker.
Conflict of interest statement
Figures

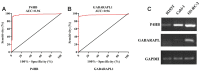
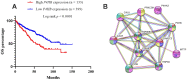
References
-
- Gremel G, Djureinovic D, Niinivirta M, Laird A, Ljungqvist O, Johannesson H, Bergman J, Edqvist PH, Navani S, Khan N, Patil T, Sivertsson Å, Uhlén M, et al.. A systematic search strategy identifies cubilin as independent prognostic marker for renal cell carcinoma. BMC Cancer. 2017; 17:9. 10.1186/s12885-016-3030-6 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

