Coolpup.py: versatile pile-up analysis of Hi-C data
- PMID: 32003791
- PMCID: PMC7214034
- DOI: 10.1093/bioinformatics/btaa073
Coolpup.py: versatile pile-up analysis of Hi-C data
Abstract
Motivation: Hi-C is currently the method of choice to investigate the global 3D organization of the genome. A major limitation of Hi-C is the sequencing depth required to robustly detect loops in the data. A popular approach used to mitigate this issue, even in single-cell Hi-C data, is genome-wide averaging (piling-up) of peaks, or other features, annotated in high-resolution datasets, to measure their prominence in less deeply sequenced data. However, current tools do not provide a computationally efficient and versatile implementation of this approach.
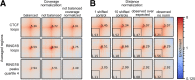
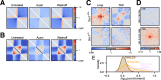
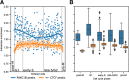
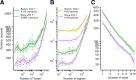
Results: Here, we describe coolpup.py-a versatile tool to perform pile-up analysis on Hi-C data. We demonstrate its utility by replicating previously published findings regarding the role of cohesin and CTCF in 3D genome organization, as well as discovering novel details of Polycomb-driven interactions. We also present a novel variation of the pile-up approach that can aid the statistical analysis of looping interactions. We anticipate that coolpup.py will aid in Hi-C data analysis by allowing easy to use, versatile and efficient generation of pile-ups.
Availability and implementation: Coolpup.py is cross-platform, open-source and free (MIT licensed) software. Source code is available from https://github.com/Phlya/coolpuppy and it can be installed from the Python Packaging Index.
© The Author(s) 2020. Published by Oxford University Press.
Figures
References
-
- Abdennur N. et al. (2018) Condensin II inactivation in interphase does not affect chromatin folding or gene expression. bioRxiv, doi: 10.1101/437459.

