Community-intrinsic properties enhance keratin degradation from bacterial consortia
- PMID: 32004342
- PMCID: PMC6994199
- DOI: 10.1371/journal.pone.0228108
Community-intrinsic properties enhance keratin degradation from bacterial consortia
Abstract
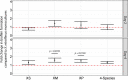
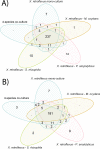
Although organic matter may accumulate sometimes (e.g. lignocellulose in peat bog), most natural biodegradation processes are completed until full mineralization. Such transformations are often achieved by the concerted action of communities of interacting microbes, involving different species each performing specific tasks. These interactions can give rise to novel "community-intrinsic" properties, through e.g. activation of so-called "silent genetic pathways" or synergistic interplay between microbial activities and functions. Here we studied the microbial community-based degradation of keratin, a recalcitrant biological material, by four soil isolates, which have previously been shown to display synergistic interactions during biofilm formation; Stenotrophomonas rhizophila, Xanthomonas retroflexus, Microbacterium oxydans and Paenibacillus amylolyticus. We observed enhanced keratin weight loss in cultures with X. retroflexus, both in dual and four-species co-cultures, as compared to expected keratin degradation by X. retroflexus alone. Additional community intrinsic properties included accelerated keratin degradation rates and increased biofilm formation on keratin particles. Comparison of secretome profiles of X. retroflexus mono-cultures to co-cultures revealed that certain proteases (e.g. serine protease S08) were significantly more abundant in mono-cultures, whereas co-cultures had an increased abundance of proteins related to maintaining the redox environment, e.g. glutathione peroxidase. Hence, one of the mechanisms related to the community intrinsic properties, leading to enhanced degradation from co-cultures, might be related to a switch from sulfitolytic to proteolytic functions between mono- and co-cultures, respectively.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation.ISME J. 2015 Jan;9(1):81-9. doi: 10.1038/ismej.2014.96. Epub 2014 Jun 17. ISME J. 2015. PMID: 24936766 Free PMC article.
-
Interspecies interactions reduce selection for a biofilm-optimized variant in a four-species biofilm model.Environ Microbiol Rep. 2019 Dec;11(6):835-839. doi: 10.1111/1758-2229.12803. Epub 2019 Nov 14. Environ Microbiol Rep. 2019. PMID: 31680421
-
Synergistic Interactions within a Multispecies Biofilm Enhance Individual Species Protection against Grazing by a Pelagic Protozoan.Front Microbiol. 2018 Jan 9;8:2649. doi: 10.3389/fmicb.2017.02649. eCollection 2017. Front Microbiol. 2018. PMID: 29375516 Free PMC article.
-
Bacterial social interactions and the emergence of community-intrinsic properties.Curr Opin Microbiol. 2018 Apr;42:104-109. doi: 10.1016/j.mib.2017.11.018. Epub 2017 Dec 1. Curr Opin Microbiol. 2018. PMID: 29197823 Review.
-
Potential use of fungal-bacterial co-cultures for the removal of organic pollutants.Crit Rev Biotechnol. 2022 May;42(3):361-383. doi: 10.1080/07388551.2021.1940831. Epub 2021 Jul 29. Crit Rev Biotechnol. 2022. PMID: 34325585 Review.
Cited by
-
Unfolding the collective functional potential of a synergistic multispecies community through genotypic and phenotypic analyses.Biofilm. 2025 May 24;10:100290. doi: 10.1016/j.bioflm.2025.100290. eCollection 2025 Dec. Biofilm. 2025. PMID: 40657038 Free PMC article.
-
Structure, Application, and Biochemistry of Microbial Keratinases.Front Microbiol. 2021 Jun 23;12:674345. doi: 10.3389/fmicb.2021.674345. eCollection 2021. Front Microbiol. 2021. PMID: 34248885 Free PMC article. Review.
-
Comparative Genomics Analysis of Keratin-Degrading Chryseobacterium Species Reveals Their Keratinolytic Potential for Secondary Metabolite Production.Microorganisms. 2021 May 12;9(5):1042. doi: 10.3390/microorganisms9051042. Microorganisms. 2021. PMID: 34066089 Free PMC article.
-
Decoding the impact of interspecies interactions on biofilm matrix components.Biofilm. 2025 Mar 14;9:100271. doi: 10.1016/j.bioflm.2025.100271. eCollection 2025 Jun. Biofilm. 2025. PMID: 40212915 Free PMC article.
-
Keratin Biomaterials in Skin Wound Healing, an Old Player in Modern Medicine: A Mini Review.Pharmaceutics. 2021 Nov 28;13(12):2029. doi: 10.3390/pharmaceutics13122029. Pharmaceutics. 2021. PMID: 34959311 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

