Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize
- PMID: 32004404
- PMCID: PMC7415785
- DOI: 10.1111/pbi.13349
Transactivation of Sus1 and Sus2 by Opaque2 is an essential supplement to sucrose synthase-mediated endosperm filling in maize
Abstract
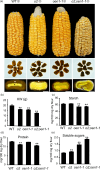
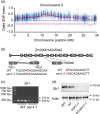
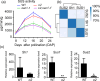
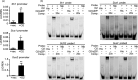
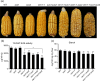
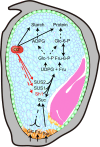
The endosperm-specific transcription factor Opaque2 (O2) acts as a central regulator for endosperm filling, but its functions have not been fully defined. Regular o2 mutants exhibit a non-vitreous phenotype, so we used its vitreous variety Quality Protein Maize to create EMS-mutagenesis mutants for screening o2 enhancers (oen). A mutant (oen1) restored non-vitreousness and produced a large cavity in the seed due to severely depleted endosperm filling. When oen1 was introgressed into inbred W64A with a normal O2 gene, the seeds appeared vitreous but had a shrunken crown. oen1 was determined to encode Shrunken1 (Sh1), a sucrose synthase (SUS, EC 2.4.1.13). Maize contains three SUS-encoding genes (Sh1, Sus1, and Sus2) with Sh1 contributing predominantly to the endosperm. We determined SUS activity and found a major and minor reduction in oen1 and o2, respectively. In o2;oen1-1, SUS activity was further decreased. We found all Sus gene promoters contain at least one O2 binding element that can be specifically recognized and be transactivated by O2. Sus1 and Sus2 promoters had a much stronger O2 transactivation than Sh1, consistent with their transcript reduction in o2 endosperm. Although sus1 and sus2 alone or in combination had no perceptible phenotype, either of them could dramatically enhance seed opacity and cavity in sh1, indicating that transactivation of Sus1 and Sus2 by O2 supplements SUS-mediated endosperm filling in maize. Our findings demonstrate that O2 transcriptionally regulates the metabolic source entry for protein and starch synthesis during endosperm filling.
Keywords: Sh1; Sus1; Sus2; O2; Quality Protein Paize; endosperm; starch; storage protein.
© 2020 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures
References
-
- Albrecht, G. and Mustroph, A. (2003) Localization of sucrose synthase in wheat roots: increased in situ activity of sucrose synthase correlates with cell wall thickening by cellulose deposition under hypoxia. Planta, 217, 252–260. - PubMed
-
- Borisjuk, L. and Rolletschek, H. (2009) The oxygen status of the developing seed. New Phytol. 182, 17–30. - PubMed
-
- Borisjuk, L. , Rolletschek, H. , Radchuk, R. , Weschke, W. , Wobus, U. and Weber, H. (2004) Seed development and differentiation: a role for metabolic regulation. Plant Biol. (Stuttg) 6, 375–386. - PubMed
-
- Carlson, S.J. , Chourey, P.S. , Helentjaris, T. and Datta, R. (2002) Gene expression studies on developing kernels of maize sucrose synthase (SuSy) mutants show evidence for a third SuSy gene. Plant Mol. Biol. 49, 15–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

