Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody
- PMID: 32004464
- PMCID: PMC7042716
- DOI: 10.1016/j.cell.2020.01.010
Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody
Abstract
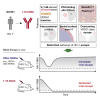
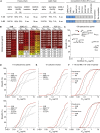
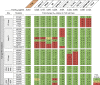
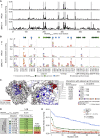
Broadly neutralizing antibodies (bNAbs) represent a promising approach to prevent and treat HIV-1 infection. However, viral escape through mutation of the HIV-1 envelope glycoprotein (Env) limits clinical applications. Here we describe 1-18, a new VH1-46-encoded CD4 binding site (CD4bs) bNAb with outstanding breadth (97%) and potency (GeoMean IC50 = 0.048 μg/mL). Notably, 1-18 is not susceptible to typical CD4bs escape mutations and effectively overcomes HIV-1 resistance to other CD4bs bNAbs. Moreover, mutational antigenic profiling uncovered restricted pathways of HIV-1 escape. Of most promise for therapeutic use, even 1-18 alone fully suppressed viremia in HIV-1-infected humanized mice without selecting for resistant viral variants. A 2.5-Å cryo-EM structure of a 1-18-BG505SOSIP.664 Env complex revealed that these characteristics are likely facilitated by a heavy-chain insertion and increased inter-protomer contacts. The ability of 1-18 to effectively restrict HIV-1 escape pathways provides a new option to successfully prevent and treat HIV-1 infection.
Keywords: CD4 binding site; HIV-1; HIV-1 escape restriction; broadly neutralizing antibodies; cryogenic electron microscopy; deep mutational scanning; escape mutations; humanized mice; immunotherapy; mutational antigenic profiling.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests A patent application encompassing aspects of this work has been filed by the University of Cologne, listing P.S., H.G., and F.K. as inventors.
Figures










References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

